Cytosine methylation (mC) is widespread in eukaryotic genomes. Most DNA methylation occurs in the CG context in animals, while DNA methylation also occurs in non-CG context (CHH and CHG) in the plants. DNA methylation is maintained by the various DNA methyltransferases. In rice, lack of non-CG methylation leads to more obvious growth and development defects than that in Arabidopsis. This suggests the essential roles of non-CG methylation in rice. However, only a few non-CG methyltransferases have been studied at present, and the regulation pattern of non-CG methylation in rice is still poorly understood.

Recently, Professor Jixian Zhai’s team from the School of Life Sciences at the Southern University of Science and Technology (SUSTech) and Research fellow Kejian Wang from the State Key Laboratory of Rice Biology at the China National Rice Research Institute, Chinese Academy of Agricultural Sciences (CNRRI, CAAS), published their research results in The Plant Cell. Their paper, entitled “Multiplex CRISPR-Cas9 editing of DNA methyltransferases in rice uncovers a class of non-CG methylation specific for GC-rich regions,” systematically analyzed the function of all non-CG DNA methyltransferases in rice and found that there was a specific GC-rich region in rice whose non-CG methyltransferases were not affected in the absence of the major non-CG methyltransferases (CMT2, CMT3, and DRM2). Furthermore, they also found that non-CG methylation in these regions could be maintained by OSCMT3b.
Using the multiplexed CRISPR-Cas9 genome-editing system, they created single- and multiple-knockout mutants for all seven non-CG DNA methyltransferases in rice. They profiled their methylation status at single-nucleotide resolution using whole-genome bisulfite sequencing (WGBS). Surprisingly, the simultaneous loss of DRM2, CMT2, and CMT3 functions, which completely erases all non-CG methylation in Arabidopsis, only partially reduced it in rice. Regions that remained heavily methylated in non-CG contexts in the rice Os-dcc (Osdrm2/cmt2/cmt3a) triple mutants are featured with high GC content. The residual non-CG methylation in the Os-dcc mutant was eliminated in the Os-ddccc (Osdrm2/drm3/cmt2/cmt3a/cmt3b) quintuple mutants, suggesting that OsCMT3b or OsDRM3 are involved in maintaining non-CG methylation in the absence of other major methyltransferases.
Furthermore, Professor Zhai’s and Kejian Wang’s teams found that OsCMT3b (Osdrm2/drm3/cmt2/cmt3a) can maintain the residual non-CG methylation of Os-dcc mutant. It suggested that the rice-specific copy of CMT3 (OsCMT3b) has gone through subfunctionalization after recent duplication and is responsible for the class of non-CG methylation at highly GC-rich regions in rice, possibly to accommodate the GC-rich context of the rice genome. These results showed that compared to Arabidopsis and several major crops, the rice genome contains a distinct cluster of non-CG methylated sites at highly GC-rich regions, whose methylation can complement the function of OsCMT3b in the Os-dcc triple mutant. In addition, the comprehensive datasets include 23 WGBS libraries, 30 mRNA libraries, and ten small RNA libraries from fifteen different mutants of rice DNA methyltransferases, thus will be a rich resource for the community to explore epigenetic regulation in monocot plants further.
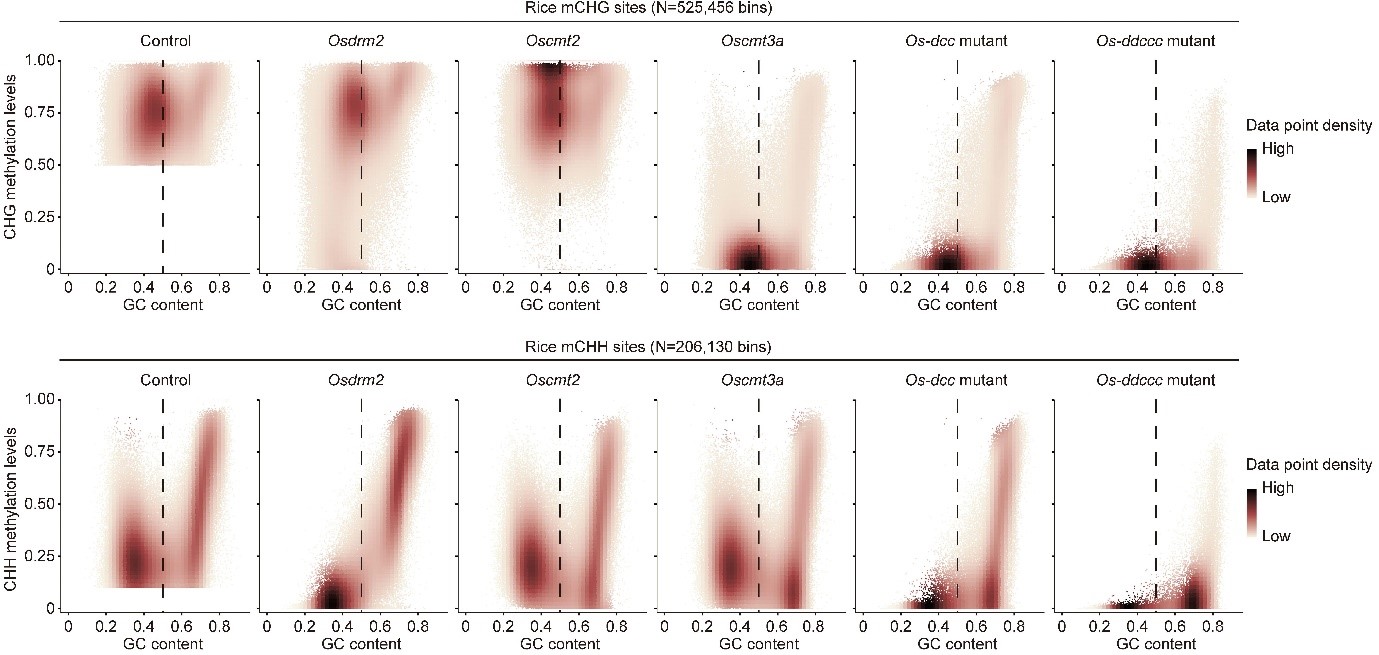
Figure 1. Average cytosine methylation levels in the CHG context of mCHG regions and CHH context of mCHH regions plotted against the GC content (CG, CHG, and CHH contexts) for different mutants.
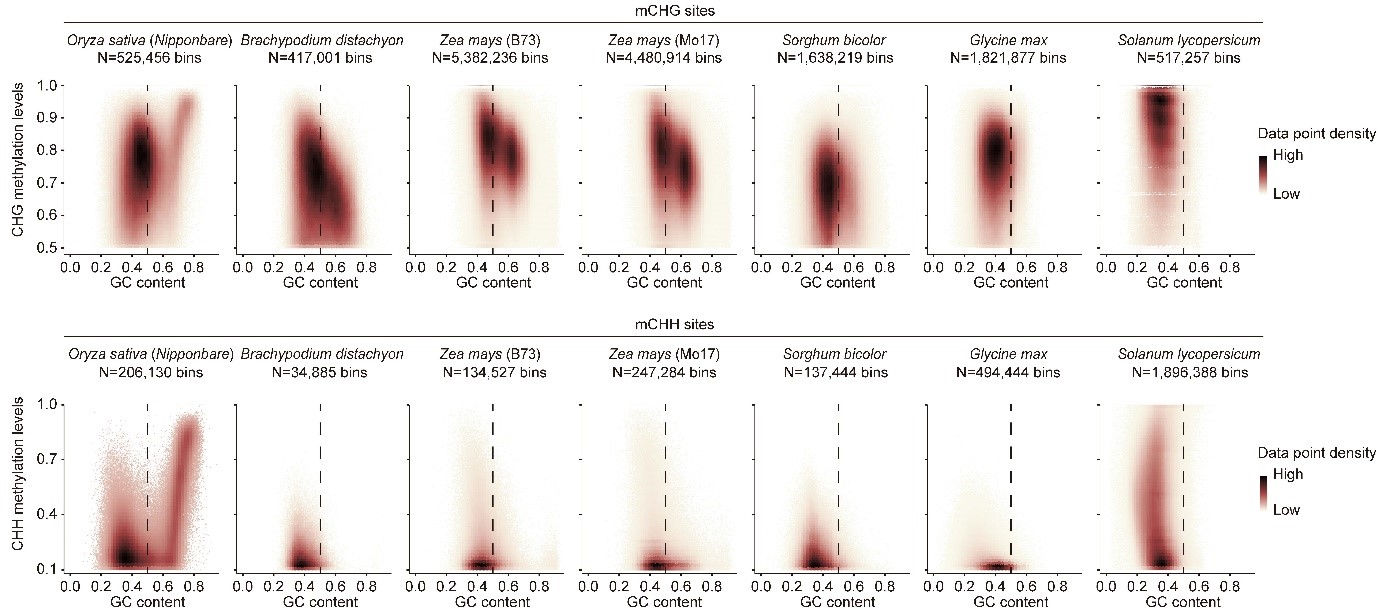
Figure 2. Average cytosine methylation levels in the CHG context of mCHG regions plotted against GC content for Brachypodium distachyon, Zea mays, Sorghum bicolor, Glycine max, and Solanum lycopersicum.
Associate Professor Jixian Zhai of the Institute of Plant and Food Science at the School of Life Sciences at SUSTech and Research fellow Kejian Wang from CNRRI, CAAS, are the co-corresponding authors of this paper. Dr. Daoheng Hu and Ph.D. student Yiming Yu of SUSTech, and Dr. Chun Wang of the CNRRI, CAAS, are the co-first authors. Professor Xuehua Zhong from the University of Wisconsin-Madison, Professor Jiamu Du from SUSTech, Professor Lei Gong from Northeast Normal University, and Professor Rui Xia from South China Agricultural University (SCAU) also made contributions to this study.
The research was supported by the National Natural Science Foundation of China (NSFC), Guangdong Innovation and Entrepreneurship Team, Shenzhen Science and Technology Innovation Commission, and the Innovation Project of the Chinese Academy of Agricultural Sciences.
Paper link: https://doi.org/10.1093/plcell/koab162
Proofread ByAdrian Cremin, Yingying XIA
Photo By