Recently, Assistant Professor Ning Jia from the School of Medicine at the Southern University of Science and Technology (SUSTech) and Dinshaw J. Patel from the Memorial Sloan Kettering Cancer Center to further examine novel mechanistic insights into the anti-CRISPR function that have emerged from X-ray crystallography and cryo-electron microscopy studies. They also looked at how these structures combined with functional studies provide valuable tools for the ever-growing CRISPR–Cas biotechnology toolbox. Their research, entitled “Structure-based functional mechanisms and biotechnology applications of anti-CRISPR proteins,” was published in Nature Reviews Molecular Cell Biology, a renowned journal in the fields of molecular and cell biology.
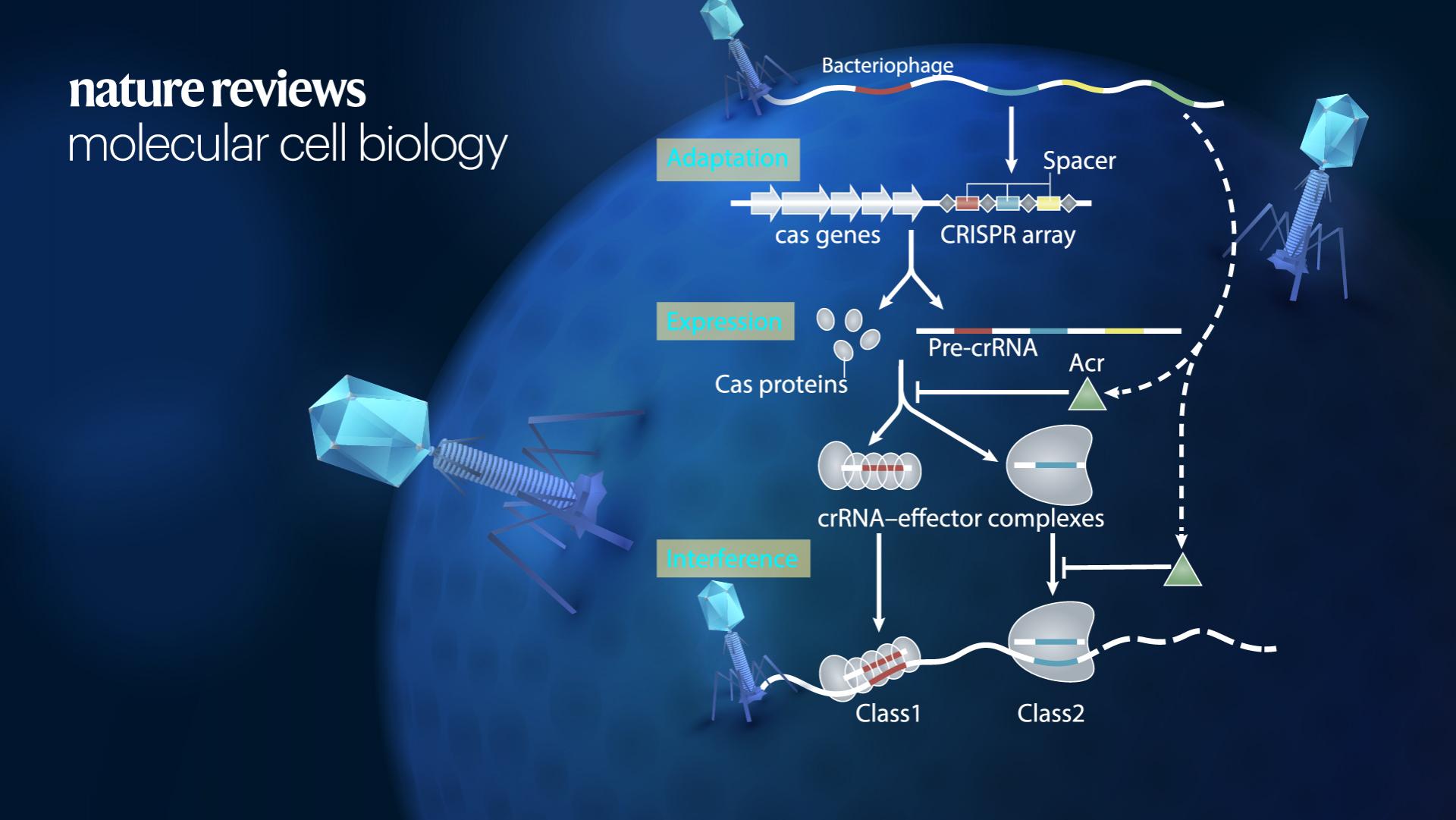
CRISPR loci and Cas proteins provide adaptive immunity in prokaryotes against invading bacteriophages and plasmids. In response, bacteriophages have evolved a broad spectrum of anti-CRISPR proteins (anti-CRISPRs) to counteract and overcome this immunity pathway. Numerous anti-CRISPRs have been identified to date, which suppresses single-subunit Cas effectors (in CRISPR class 2, type II, V, and VI systems) and multisubunit Cascade effectors (in CRISPR class 1, type I and III systems).
Crystallography and cryo-electron microscopy structural studies of anti-CRISPRs bound to effector complexes, complemented by functional experiments in vitro and in vivo, have identified four major CRISPR–Cas suppression mechanisms. These were inhibition of CRISPR–Cas complex assembly, blocking target binding, prevention of target cleavage, and degradation of cyclic oligonucleotide signaling molecules.
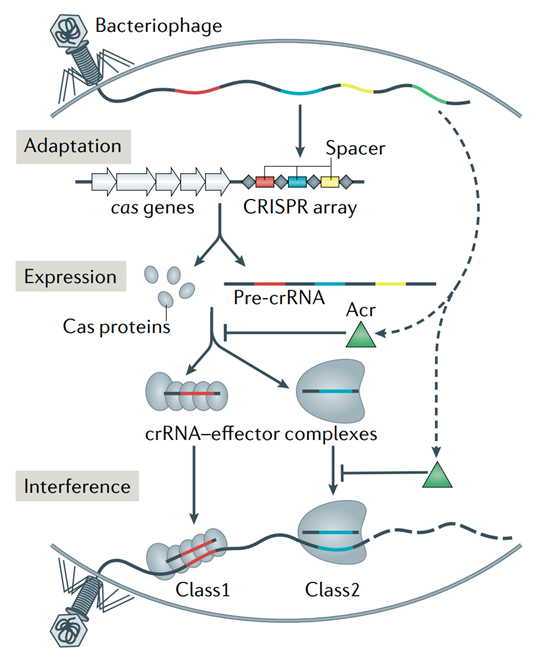
Figure 1. Box 1 CRISPR–Cas adaptive immunity in prokaryotes
Assistant Professor Ning Jia from the School of Medicine at SUSTech is the first author of this paper. Ning Jia and Dinshaw J. Patel from the Memorial Sloan Kettering Cancer Center are the co-corresponding authors.
Paper link: https://www.nature.com/articles/s41580-021-00371-9
Proofread ByAdrian Cremin, Yingying XIA
Photo By