High-throughput RNA-sequencing (RNA-seq) has become the most popular technology for profiling gene expression in the last decade due to its low cost and high coverage. As a result, the number of RNA-seq libraries from the plant community has increased exponentially in recent years. For major crops, such as maize, rice, soybean, wheat, and cotton, the plant community has collected a total of ~45,000 libraries by 2021.
However, these existing databases only host the already processed data from each study separately. Therefore, the expression values cannot be directly compared among projects because they were derived from different bioinformatic pipelines and often mapped to different versions of the reference genomes. To take full advantage of the big data of RNA-seq libraries, an effort to integrate all publicly available libraries via a uniformed processing pipeline and curate them into an easy-to-use searchable database is urgently needed.
Associate Professor Jixian Zhai’s group from the Institute of Plant and Food Science of the School of Life Sciences at the Southern University of Science and Technology (SUSTech) recently released a comprehensive online database that allows easy and fast access to ~45,000 plant public RNA-seq libraries. This study, entitled “PPRD: a comprehensive online database for expression analysis of ~45,000 plant public RNA-Seq libraries,” has been published in Plant Biotechnology Journal, a high-impact journal with an emphasis on molecular plant sciences and their applications through plant biotechnology.
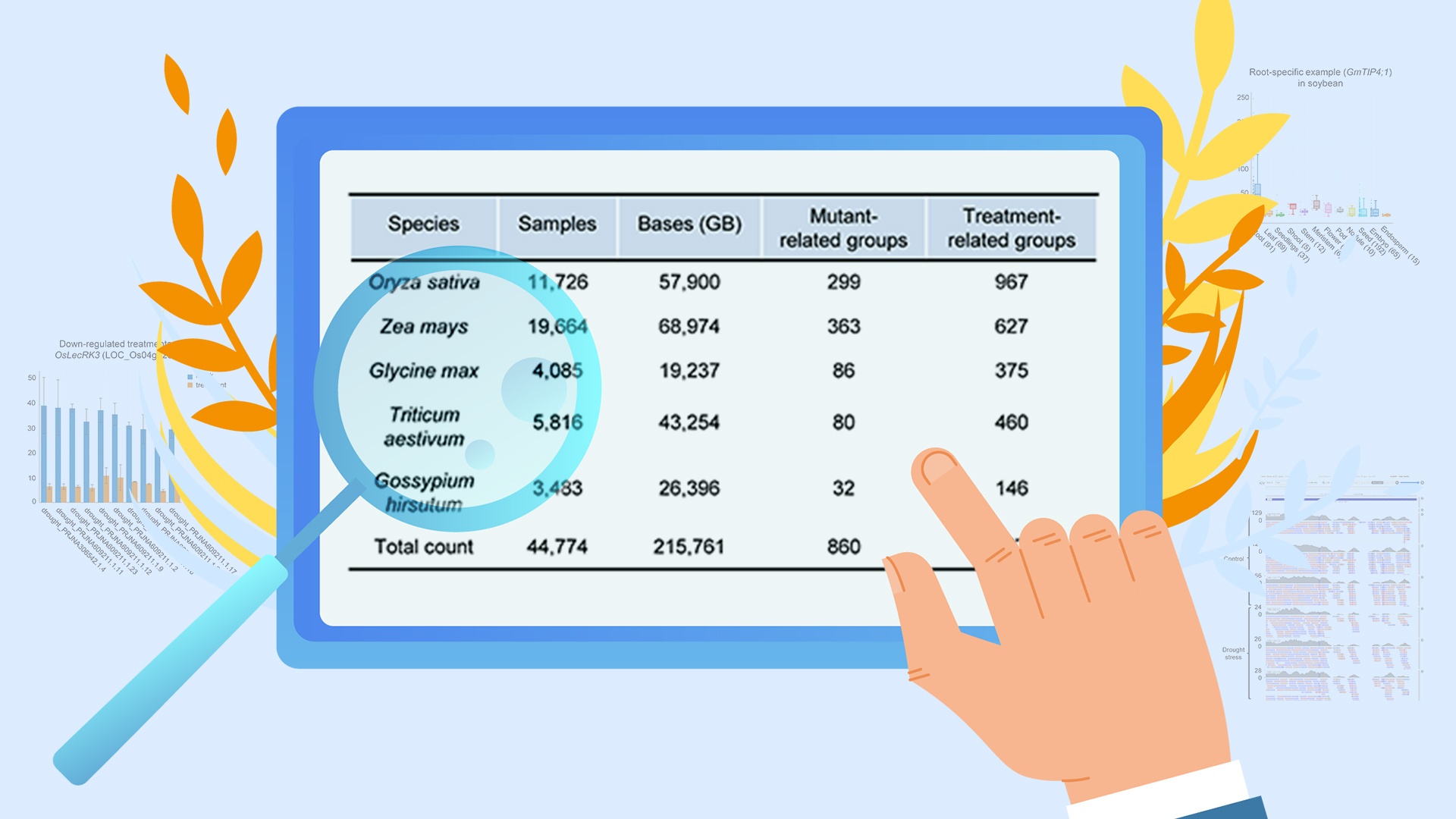
To address this challenge, the researchers proposed a comprehensive web-based platform, Plant Public RNA-seq Database (PPRD). PPRD consists of a large number of RNA-seq libraries of maize (19,664), rice (11,726), soybean (4,085), wheat (5,816), and cotton (3,483) from Gene Expression Omnibus (GEO), Sequence Read Archive (SRA), European Nucleotide Archive (ENA), and DNA Data Bank of Japan (DDBJ) databases (Figure 1B). These RNA-seq data are manually curated to highlight different mutants, tissues, developmental stages, abiotic or biotic stresses. Besides showing expression patterns from different tissues and developmental stages (Figure 1C-1E), they also annotated the mutant-related groups and treatment-associated groups in the maize, rice, soybean, wheat, and cotton database, respectively.
In general, PPRD supports searches by gene ID, library ID, BioProject IDs, keywords, or any combination of these terms in selected libraries. After querying the above terms, the results in tables and diagrams were returned, including the results of expression comparison in multiple interactive diagrams, expression levels among different tissues, developmental stages, abiotic and biotic stresses, and up-regulated or down-regulated expression in mutant-related or treatment-related samples. The “CoExpression” page provides a list of genes co-expressed with the searched one. The “IGV Online” page is flexible for visualizing the mapping landscape of the local genomic region in selected libraries. In addition, the “Share” function was supported to facilitate showing the results with others.
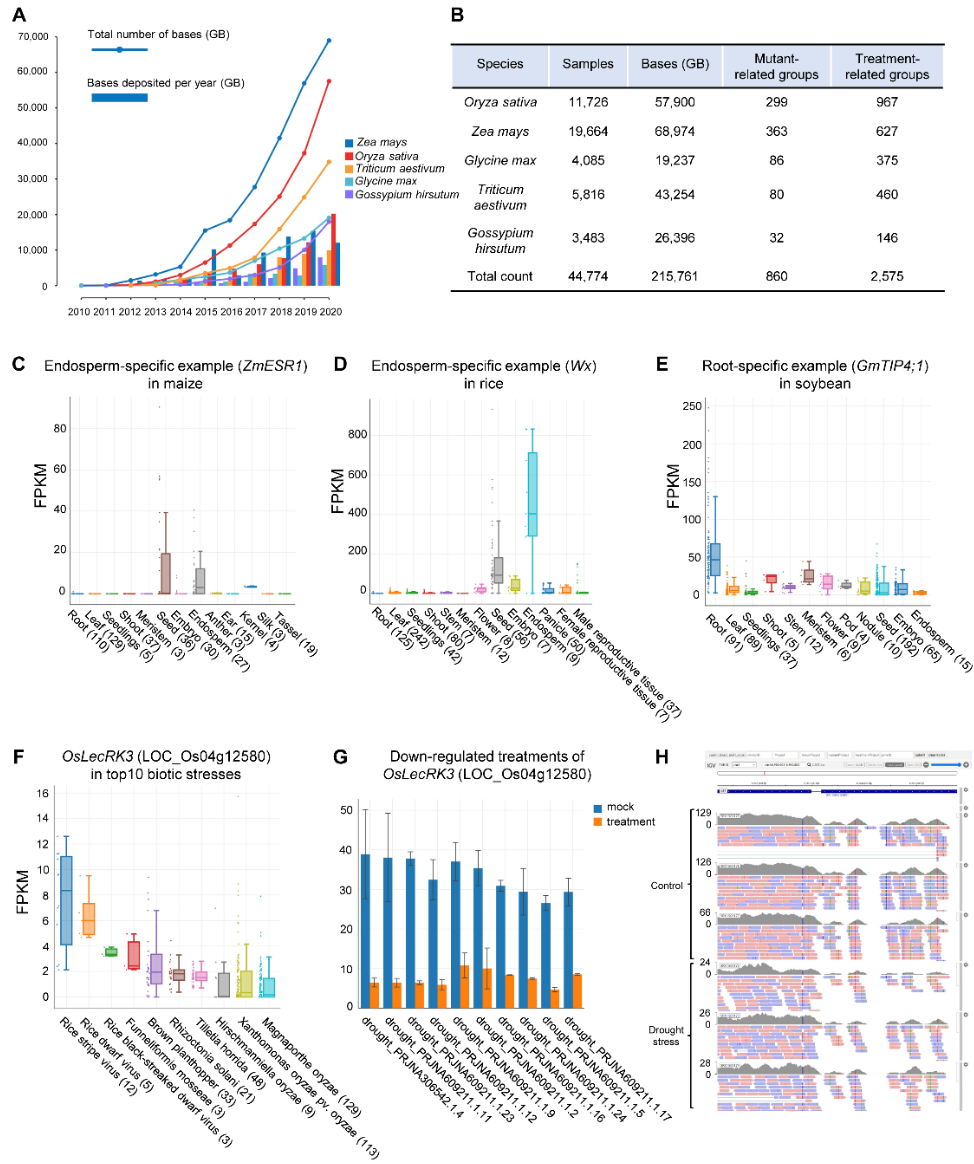
Figure 1. Overview of Plant Public RNA-Seq Database. (A) The number of Oryza sativa, Zea mays, Glycine max, Triticum aestivum, and Gossypium hirsutum sequenced bases per year from 2010 to 2020. The bar indicates the bases deposited per year (GB). The line indicates the total number of bases (GB). GB: Giga base pairs. (B) The basic summary of RNA-seq libraries. “Mutant-related groups” and “Treatment-related groups” denote the number of groups used to analyze the differential expression. (C-E) The tissue-specific expression of some marker genes. The left panel shows the endosperm-specific expression of ZmESR1 in maize (C), the middle panel shows the endosperm-specific expression of Wx in rice (D), and the right panel displays the root-specific expression of GmTIP4;1 in soybean (E). (F) The expression level of OsLecRK3 (LOC_Os04g12580) among top10 biotic stresses in rice. (G) Down-regulated expression of OsLecRK3 (LOC_Os04g12580) among top10 treatment groups in rice. (H) The overview of IGV. The mapped reads of OsLecRK3 show decreased abundance in drought stress-related samples.
Yiming Yu, a Ph.D. student, and Hong Zhang, a master’s student of Associate Prof. Jixian Zhai’s group at SUSTech, are the co-first authors of this paper. Associate Prof. Zhai is the co-corresponding author. Research Assistant Professor Yanping Long and Ph.D. student Yi Shu also made contributions to this study. The research was supported by the Guangdong Innovation and Entrepreneurship Team.
Related links:
Paper link: https://onlinelibrary.wiley.com/doi/10.1111/pbi.13798
Plant Public RNA-seq Database (PPRD): http://ipf.sustech.edu.cn/pub/plantrna/
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By