The nucleus, the hallmark of eukaryotes distinguished from prokaryotes, is enclosed by a bilayer membrane (nuclear membrane, NE). The presence of NE hinders the cytoplasmic transportation of macromolecules. Embedded in NE, nuclear pore complexes (NPCs) are the massive bidirectional transport channels between the nucleus and the cytoplasm, and their dysfunction can cause many serious diseases, including cancer. Since the first glimpse about seventy years ago, NPCs have attracted increasing attention from researchers. However, due to the intrinsic structural flexibility, complicated composition, and huge size, the accurate molecular arrangement of NPC subunits remains thus far enigmatic.
Professor Sen-Fang Sui’s team from the School of Life Sciences and the Cryo-Electron Microscopy Center at the Southern University of Science and Technology (SUSTech) recently reported the near-atomic structure of the inner ring of the nuclear pore complex (NPC). They revealed the detailed molecular mechanism underlying the conformational change between “dilation” and “constriction” of NPC. It has provided a theoretical basis for further understanding NPC’s adaptation to different physiological environments.
Their research, entitled “Near-atomic Structure of the Inner Ring of the Saccharomyces cerevisiae nuclear pore complex,” was published in Cell Research, a journal covering all areas of life sciences.

The NPC is the largest proteinaceous assembly in vivo, which consists of ~550-1000 molecules of ~30 different nucleoporins (Nups) with the molecular mass of 60–120 MDa from yeast to human. The NPC presents a canonical sandwich architectural feature and consists of four coaxial ring structures: two outer rings on the cytoplasmic side (cytoplasmic ring, CR) and nuclear side (nuclear ring, NR) respectively, an inner ring (IR) between two layers of outer rings, and a luminal ring (LR) inserted inside the double-layer NE. As the most conserved module of NPC and connected to the other three rings, IR exhibits C8 symmetry, and one symmetric unit is named IR monomer. Furthermore, the IR monomer has a C2 (pseudo) symmetry and could be divided into two approximately the same IR protomers.
In this study, using cryo-electron microscopy together with methods of biochemistry and cell biology, the researchers successfully resolved the structures of IR monomer (3.73 Å), IR protomer (3.71 Å), and several important subunits. Finally, they built the most detailed and accurate atomic model of the intact IR so far (Figure 1).
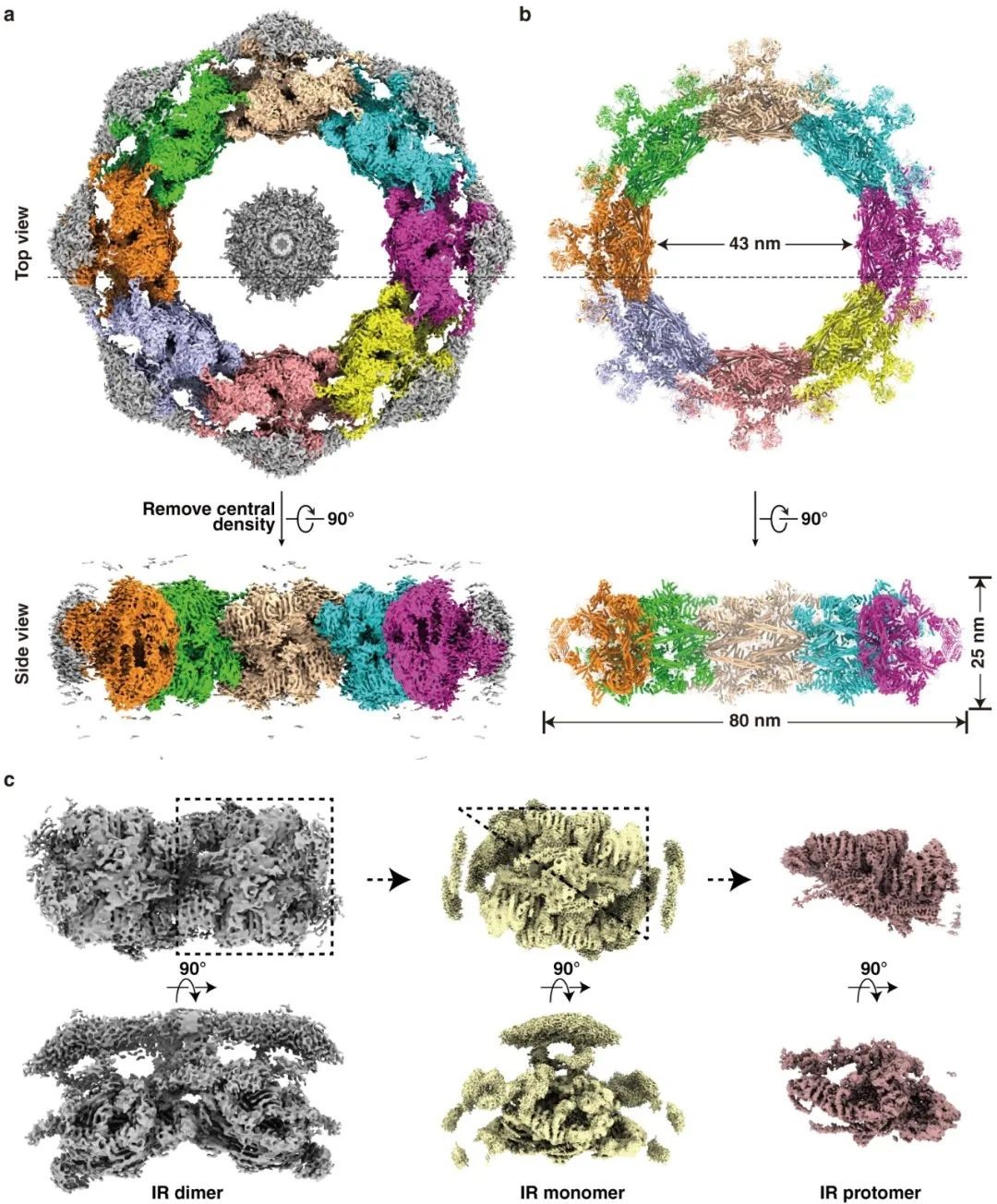
Figure. 1 Cryo-EM structure of the IR of the Saccharomyces cerevisiae NPC. (a-b) Cryo-EM density map (a) and the atomic model (b) of the intact IR. (c) Density maps of IR dimer, IR monomer, and IR protomer determined in the study.
The model of intact IR includes 192 protein molecules with a mass weight of ~16 MDa, accounting for approximately 1/3 of the entire NPC. IR monomer can be further divided into three layers: the outer, middle, and inner layers. The N-terminal ends of Nup157 and Nup170 from the outer layer protrude into the NE, indicating that Nup157 and Nup170 participate in the conjunction between IR and LR. The “Z” shaped Nup188 and Nup192 from the middle layer forms an arched cavity to accommodate the rhombic CNT tetramer from the inner layer. Each CNT tetramer contains eight hinge regions internally, which are unstable and prone to change, and eight CNT tetramers form the innermost transport channel of NPC. Therefore, it is reasonable to deduce that the structural variability of the CNT tetramer could be an adaptation to the passage of cargoes of different sizes by their own structural change.
The structural analysis showed that extensive interactions with large interfaces exist within IR monomers in contrast to the weak interactions between IR monomers. Compared with the previously published in situ structure, the structure of NPC obtained in this study was found to be a “constriction” state, and the adjacent IR monomers were associated with the above weak interactions. In the in-situ structure of “dilation”, these interactions were separated to create a nearly 7 nm “gap”. At the same time, the IR monomer, as a whole, remains relatively stable during the movement due to the presence of a host of the inner large-interfaced interactions (Figure 2).
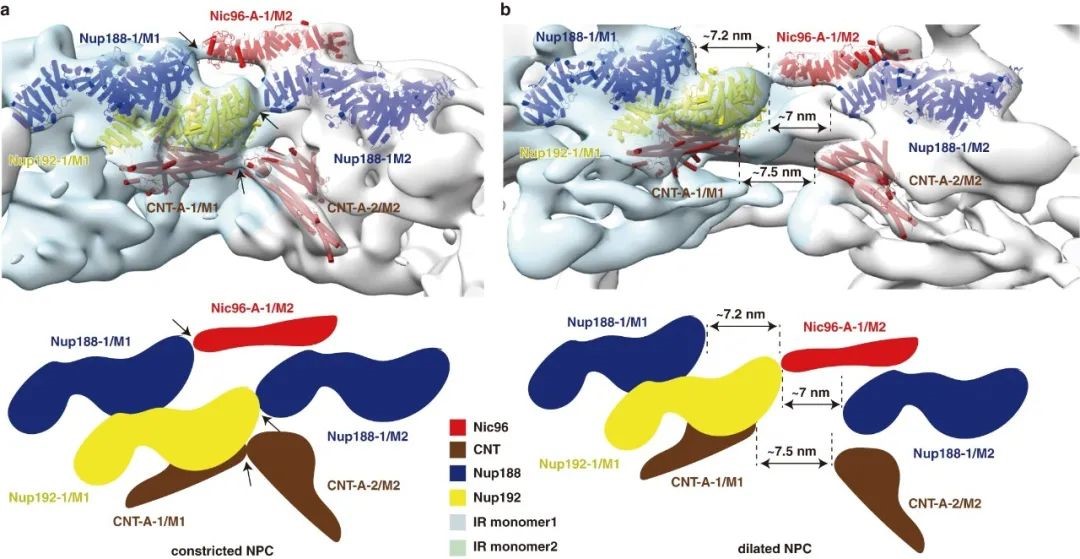
Figure. 2 Comparison of dilated and constricted NPC. (a-b) Distance changes between IR monomers are shown from constricted (a) to dilated (b) states. Schematic diagrams are drawn at the bottom, and the representative subunits of IR are color coded.
These results provide a structural basis for the in-depth understanding of NPC’s hierarchical assembly and conformational changes and their powerful physiological roles. They are of great significance for understanding the pathogenesis of related diseases and developing corresponding clinical treatment and specific drugs.
Zongqiang Li, Shuaijiabin Chen, Guoqiang Huang, all Ph.D. students from Tsinghua University and visiting students from SUSTech, alongside Liang Zhao, a postdoctoral from Tsinghua University and a visiting scholar from SUSTech, are the co-first authors of this paper. Professor Sen-Fang Sui is the corresponding author. Associate Prof. Shan Sun from Tsinghua University and Prof. Peiyi Wang from the Cryo-Electron Microscopy Center at SUSTech also participated in this research.
The data collection and image processing of this study was supported by the Cryo-Electron Microscopy Center at SUSTech.
Paper link: http://www.nature.com/articles/s41422-022-00632-y
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By