Under appropriate conditions, seeds germinate and develop into seedlings within a few days, representing a major developmental switch for flowering plants. Like other developmental progressions, germination involves dramatic alterations of gene expression, but it is challenging to distinguish between the primary causes and secondary consequences of germination. Control of gene expression, especially the stable repression of transcription, often requires chromatin level regulation, which is poorly understood for seed germination. The phytohormone (ABA) represses germination. ABA biosynthesis is restricted in seeds that are capable to germinate, but whether this is a regulated process or a default/pre-set state is unclear.
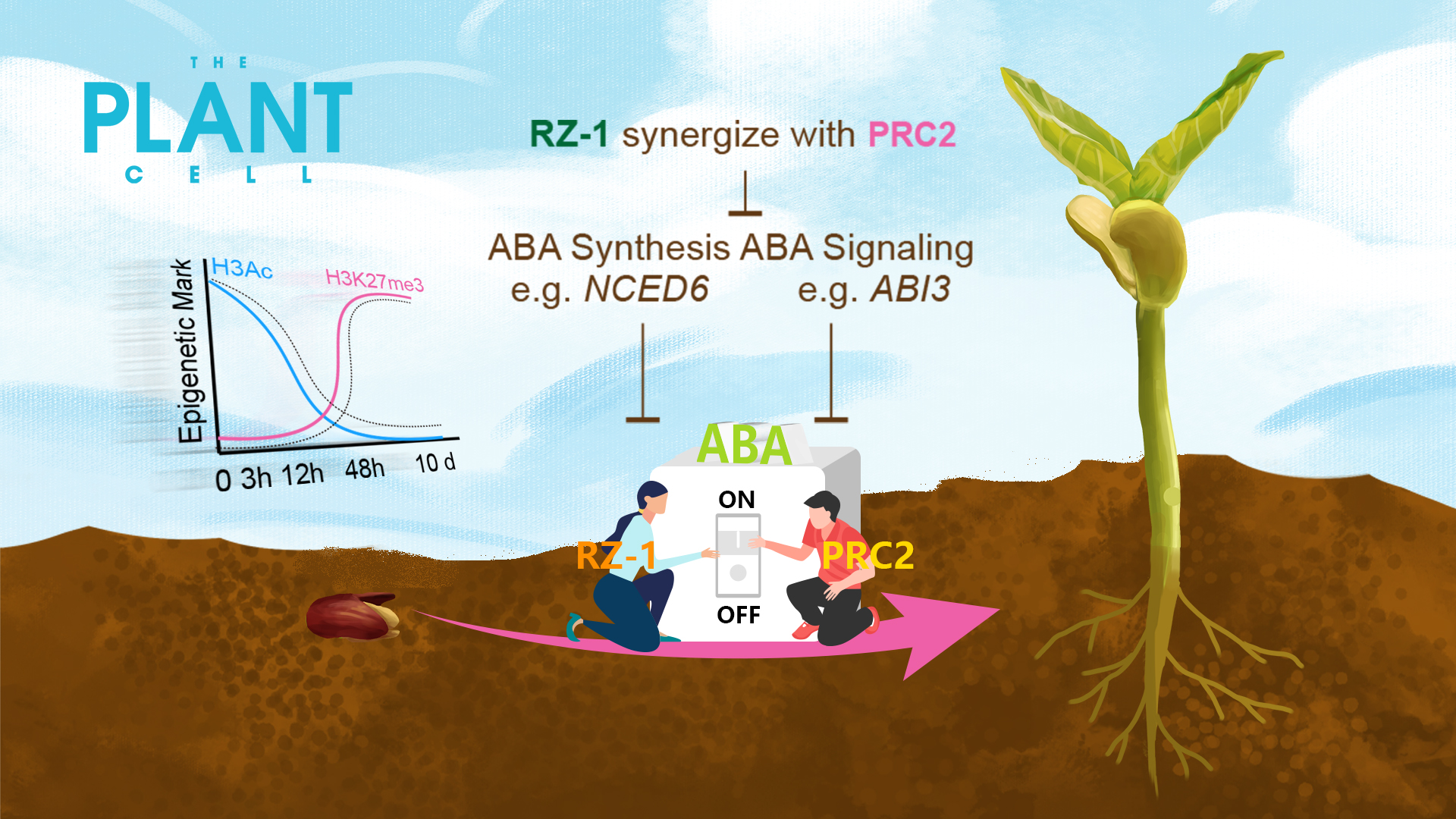
Assistant Professor Zhe Wu’s group from the School of Life Sciences at the Southern University of Science and Technology (SUSTech) has recently discovered a new regulatory mechanism for seed germination, involving synergistic interaction between an RNA binding protein RZ-1 and polycomb repressive complex 2 (PRC2) for ABA biosynthesis gene silencing.
This work, entitled “Progressive chromatin silencing of ABA biosynthesis gene permits seed germination in Arabidopsis,” has been published in The Plant Cell, a high-impact journal in the plant field.
Previous work by Prof. Zhe Wu has shown that loss of function of both RZ-1B and RZ-1C (RZ-1), a homologous pair of RNA-binding proteins with redundant functions, delays germination. Using a series of phytohormone studies and gene expression analysis, members from Prof. Wu’s lab first discovered that ABA synthesis or signaling pathway is induced in rz-1b rz-1c double mutant during seed germination. The following genetic experiment results suggested that NCED6, a gene encoding a key enzyme in ABA biosynthesis, is responsible for the delayed germination phenotype of rz-1b rz-1c mutant (Figure 1).
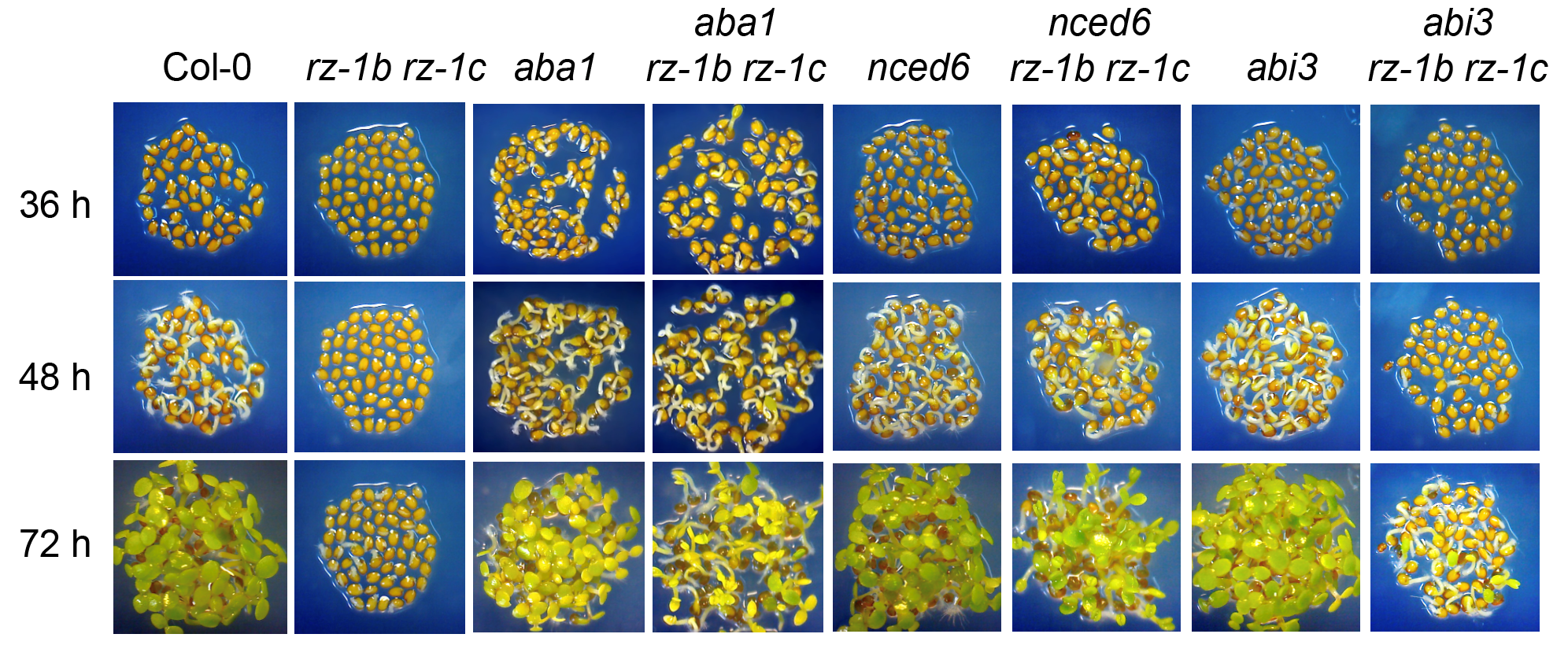
Figure 1. Genetic crossing with ABA mutants recovers germination of rz-1b rz-1c
Next, the team continued asking how RZ1, as an RNA binding protein, might work to repress the transcription of genes such as NCED6. Using the RIP-qPCR technique in different cell fractions, they found that RZ1 binds to nascent RNAs of target genes at the chromatin level. Considering PRC2 is famous for chromatin silencing, and that genes like NCED6 and ABI3 were shown to have H3K27me3 modification in seedlings, it is possible that RZ1 is also involved in the same silencing process. Indeed, PRC2 core mutant swn clf exhibits similar germination and molecular phenotype as rz-1b rz-1c; quadruple mutant swn clf rz-1b rz-1c fails to germinate with exaggerated gene expression change. These suggest that RZ1 and PRC2 act in synergy to repress specific genes during germination (Figure 2).

Figure 2. RZ-1 and PRC2 synergistically regulate seed germination and gene expression
The researchers used proteomics techniques to identify RZ-1C-associated proteins in germinating seeds, including various histone modification-related proteins, such as HDA19, AL, MSI1, and DDB1-CLU4. Further epigenomic analyses using ChIP-seq revealed altered dynamics of H3Ac, H3K4me3, and H3K27me3 in rz-1b rz-1c compared to WT. Notably, both active marker removal and repressive marker establishment are delayed in rz-1b rz-1c, consistent with induced gene expression in the mutant (Figure 3). These support the idea that RZ-1 is involved in transcription repression at the chromatin level.
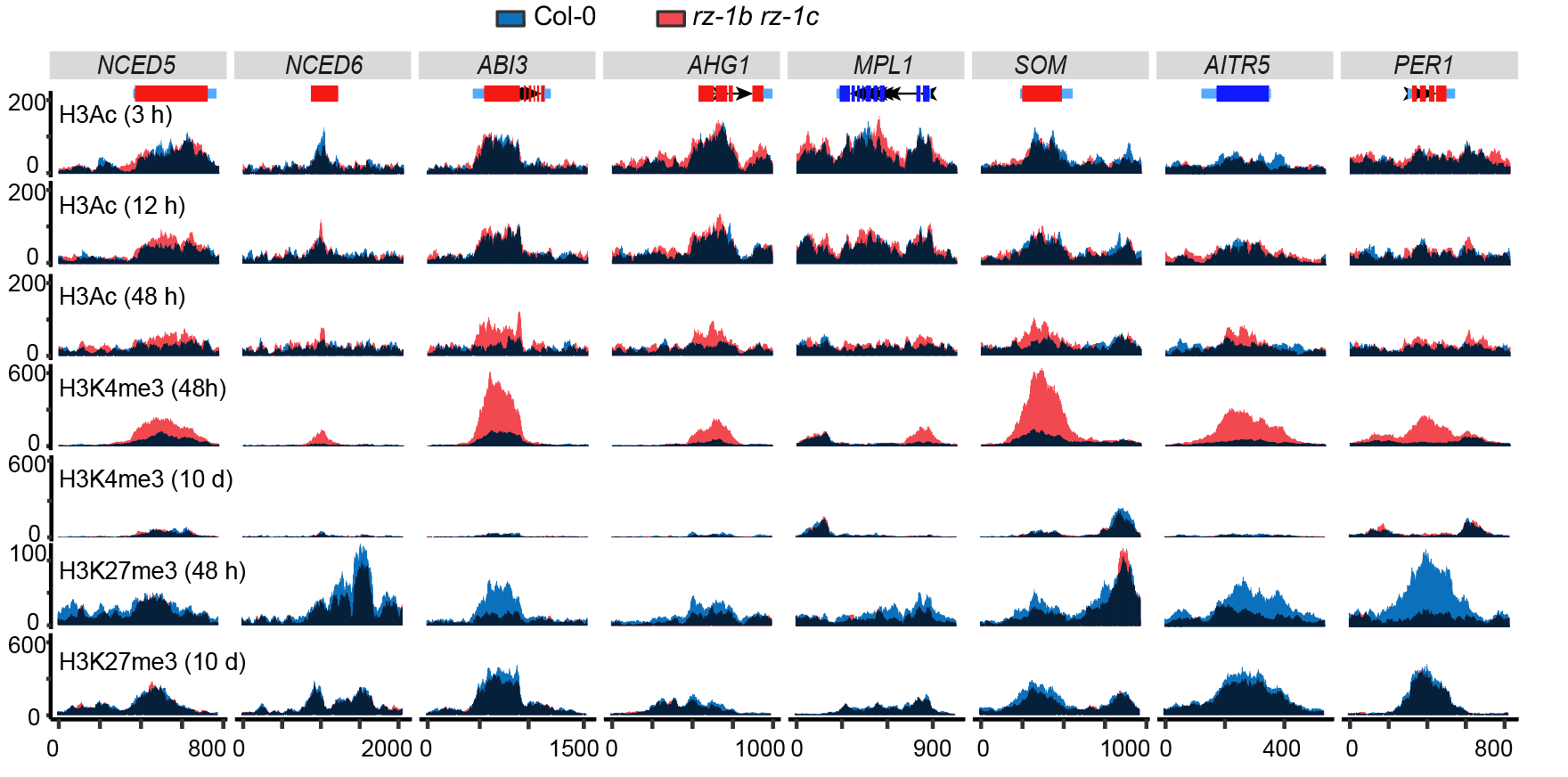
Figure 3. RZ-1 participates in histone modification change at genes to be silenced during germination
During seed germination, NCED6 and ABI3 are progressively silenced at the transcriptional and chromatin levels. This requires the coordinated action of RZ-1 and PRC2, as germination is effectively blocked when both these proteins are disabled. In early imbibition, RZ-1 targets NCED6 and ABI3, and triggers transcriptional repression, involving histone deacetylation and suppression of H3K4me3. This process facilitates and is followed by the establishment of H3K27me3, which mainly occurs at the onset of the seed-to-seedling transition. Therefore, among the thousands of genes that undergo altered expression during germination, the progressive silencing of NCED6 is a key trigger for germination (Figure 4).
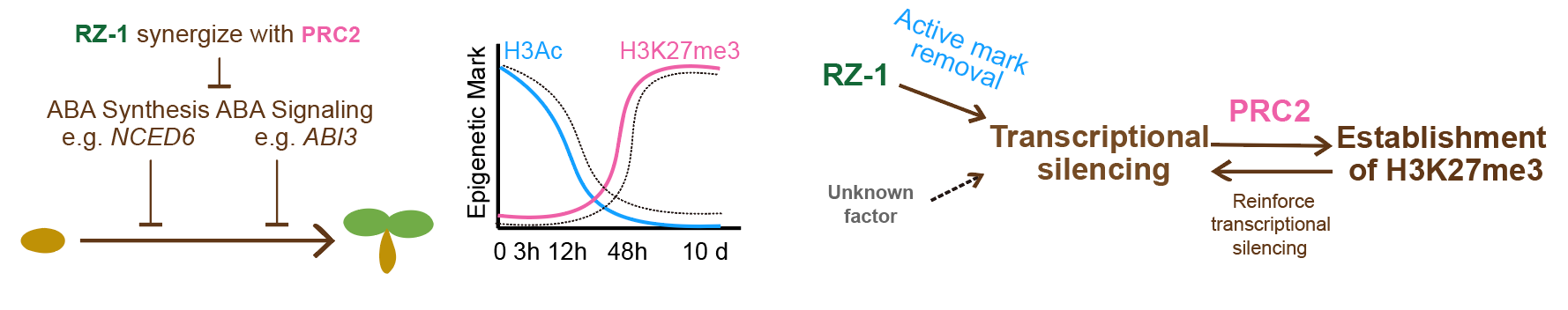
Figure 4. Proposed working mechanism for RZ1 and PRC2 in germination regulation
Deyue Yang, a senior research fellow, and Fengli Zhao, a postdoctoral fellow from Dr. Wu’s group at SUSTech, are the co-first authors of this paper. Asst. Prof. Zhe Wu is the corresponding author. Prof. Xi Chen and Prof. Jiamu Du at SUSTech, Prof. Lijia Qu at Peking University, Prof. Yufeng Wu at Nanjing Agriculture University, and Prof. Min Chen at Henan University also contributed to this study.
This work was also supported by Dr. Jinfeng Qi and Dr. Jianqiang Wu of the Kunming Institute of Botany, Chinese Academy of Science (KIB, CAS), Dr. Caroline Dean of the John Innes Centre, UK (JIC), Dr. Hongwei Guo at SUSTech, Dr. Yong Xiang of the Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences (AGIS, CAAS), and Dr. Jigang Li at China Agricultural University.
This work was funded by the National Natural Science Foundation of China (NSFC), Guangdong Innovation Research Team Fund, the Shenzhen Sci-Tech Fund, and Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes.
Paper link: https://doi.org/10.1093/plcell/koac134
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By