SARS-CoV-2 keeps evolving into new variants due to sustained global transmission. Variants with evidence of increased transmissibility, severe disease, reduction in neutralization by antibodies of convalescents or vaccines, and posing a higher risk of eluding testing are classified as Variants of Concern (VOC). A recent variant named Omicron, which obtains a large number of mutations in the receptor-binding domain (RBD) of the spike protein, has risen to intense scientific and public attention.
Professor Peiyi Wang from the Department of Biology and Cryo-Electron Microscopy Center at the Southern University of Science and Technology (SUSTech), in collaboration with researchers from the Institute of Microbiology, Chinese Academy of Sciences (IMCAS), reported the complex structure of human ACE2 to spike RBD from Omicron and Delta SARS-CoV-2 and revealed the molecular mechanism.
These results, entitled “Receptor binding and complex structures of human ACE2 to spike RBD from Omicron and Delta SARS-CoV-2”, have been published in Cell.
Currently, five VOCs have been announced by the World Health Organization (WHO). Omicron and Delta are the two most dangerous variants of the five VOCs. Omicron has appeared in 128 countries and thus attracted worldwide attention. The RBD of the spike protein of Omicron carries up to 15 amino acid mutations, covering all the characteristics of the Alpha, Beta, and Gamma variants. The Delta variant is the most transmissible COVID-19 variant found so far. An in-depth understanding of the receptor recognition and cell invasion mechanism of Omicron and Delta mutants is the key foundation of vaccine and drug research and development.
The researchers measured the binding affinities of the five VOCs to hACE2 and pseudo virus infections through qualitative and quantitative research methods such as flow cytometry analysis, surface plasmon resonance (SPR), and pseudo virus invasion experiments. The results demonstrated that Omicron along with the Delta variant RBD binds to hACE2 with comparable affinity to that of the prototype.
To further characterize the binding modes of hACE2 with Omicron RBD or Delta RBD, the researchers used both X-ray crystallography and Cryo-electron microscopy (cryo-EM) to determine the complex structures. The crystal and cryo-EM structures of hACE2 with Omicron RBD were resolved at a resolution of 3.0 Å and 3.4 Å, respectively. The crystal structure of the hACE2/Delta RBD complex was also determined at a resolution of 3.35 Å (Figure 1).
Structural analysis shows that K417N, G446S, E484A, G496S, and Y505H substitutions decrease the binding affinity of Omicron RBD with hACE2, whereas Y41 from hACE2 with π-π stacking interaction with Y501 of RBD compensatively increases the binding affinity of Omicron RBD with hACE2. In addition, two mutation sites on Delta RBD, L452R, and T478K do not affect its binding to hACE2, but may affect the binding of antibodies resulting in immune escape when treated with most clinically available monoclonal antibodies.
This study revealed the molecular mechanism of the interaction between RBD and hACE2 of the two most concerned COVID-19 variants Omicron and Delta, which laid a molecular foundation for vaccine research and drug screening.
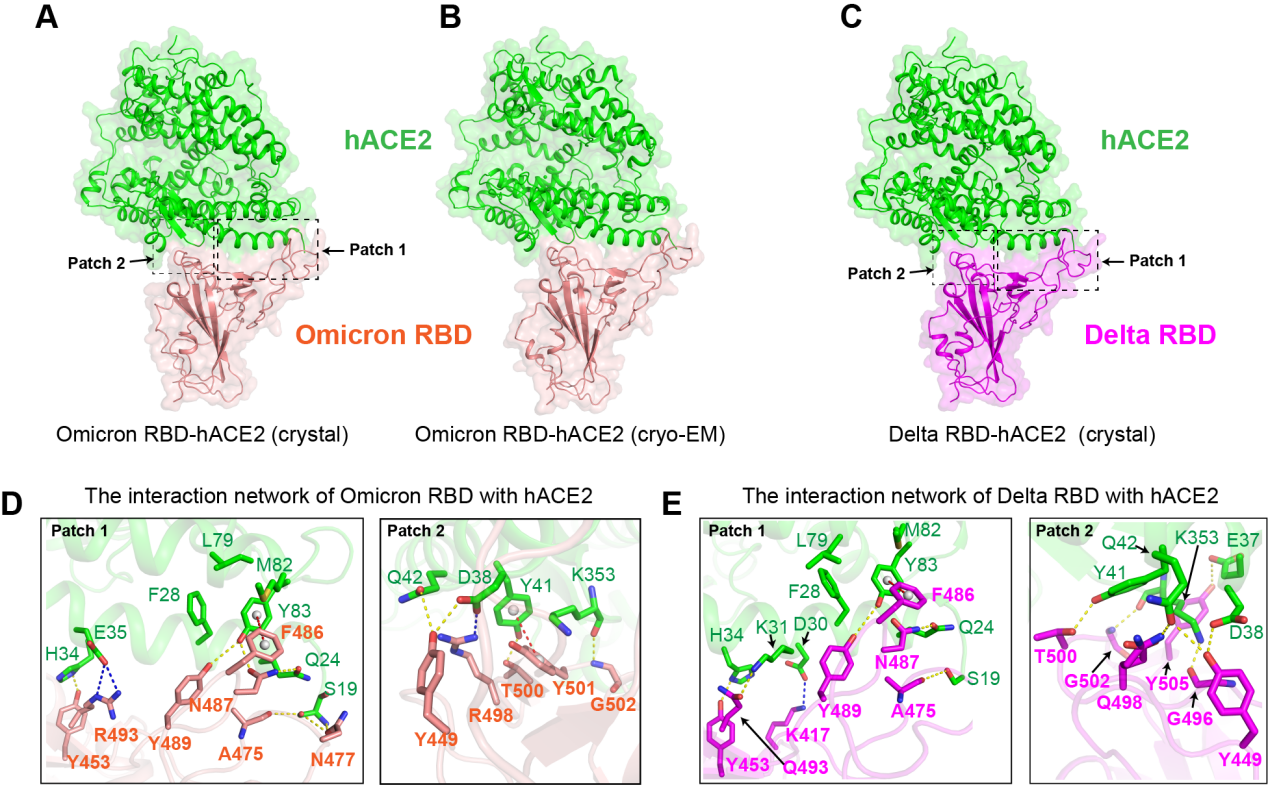
Figure 1. The Omicron and Delta RBD-hACE2 complex structures
Sheng Liu from SUSTech, Pengcheng Han and Linjie Li from IMCAS, Qisheng Wang from the Shanghai Synchrotron Radiation Facility (SSRF), and Di Zhang from the Chinese Academy of Sciences (CAS) are the first authors of this paper. Prof. Peiyi Wang from SUSTech and Profs. George F. Gao and Jianxun Qi from IMCAS are the corresponding authors.
This work was supported by the National Key R&D Program of China, the Shenzhen Science and Technology Program, and the SUSTech Presidential Postdoctoral Fellowship Program. Cryo-EM image data collection and image processing were supported by the Cryo-Electron Microscopy Center of SUSTech.
Paper link: https://doi.org/10.1016/j.cell.2022.01.001
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By