The ongoing emergence of SARS-CoV-2 Omicron variants continues to pose a significant health threat due to their enhanced transmissibility and immune evasion capabilities. Among these Omicron variants, the BA.2 variant and its derived subvariants, such as XBB.1 and BA.2.75, have gained prominence for their high infectivity and immune evasion potential. BA.2 variant diversified into multiple sub-lineages. One branch gave rise to the BA.4 and BA.5 sub-lineages, which share the same Spike (S) protein. Compared to the original strain, the Omicron sub-lineages harbor more than 30 mutations in the S protein. The evolution of glycan moieties on the S protein remains an elusive aspect requiring further investigation.
Assistant Professor Renhong Yan’s and Associate Professor Liwei Cao’s research groups from the School of Medicine at the Southern University of Science and Technology (SUSTech) have recently published their latest cooperative research results where they have identified a convergent binding pattern between RBD and ACE2 in the BA.2.75, BF.7, and XBB.1 subvariants, suggesting ACE2 mimic monoclonal antibodies might have the most broad-spectrum neutralizing potency.
Their work, entitled “Cryo-EM structures of SARS-CoV-2 BA.2 derived subvariants spike in complex with ACE2”, has been published in Cell Discovery.
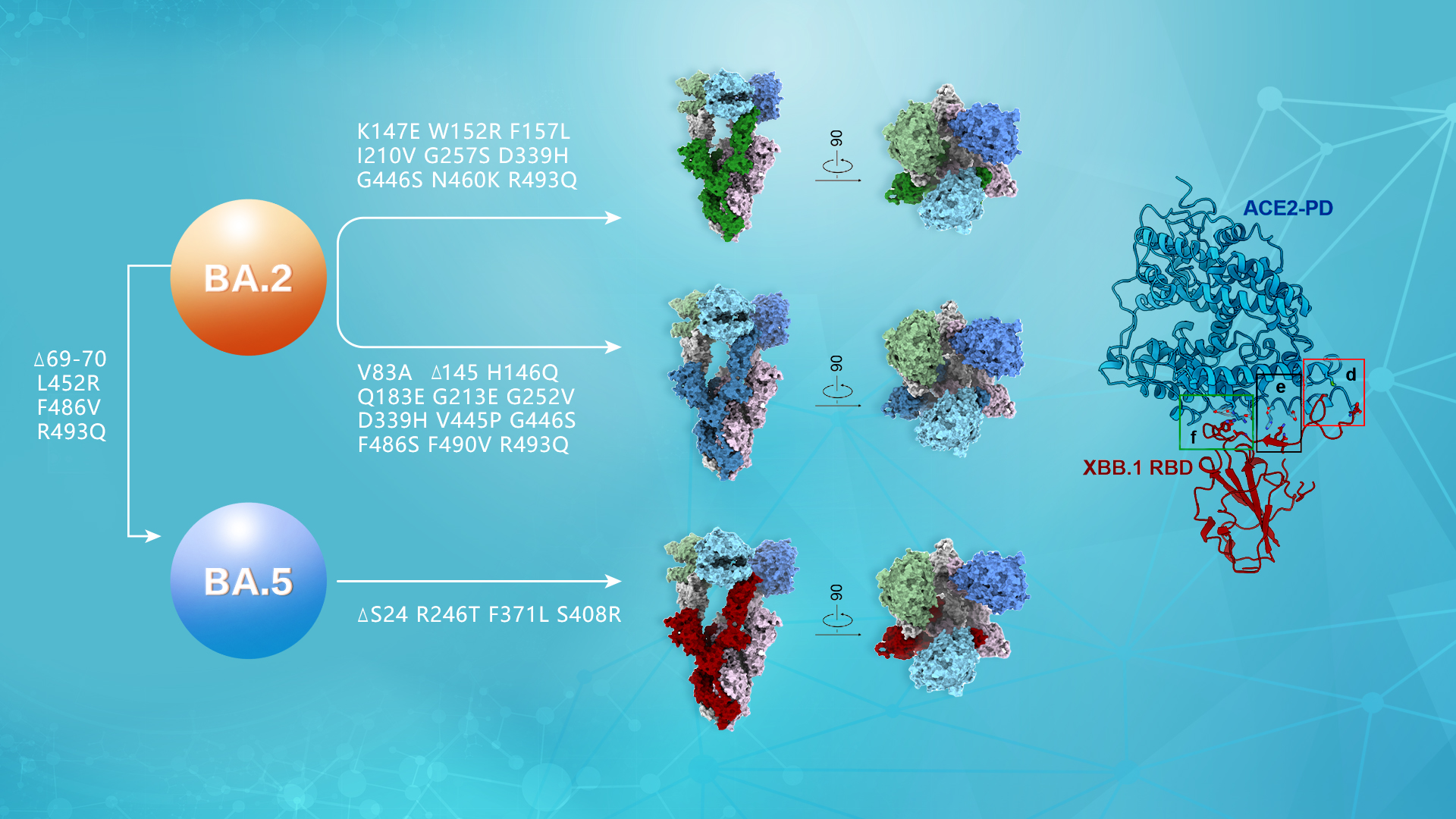
To elucidate the biochemical properties of the S protein in Omicron mutant strains, the researchers measured the affinities of the S proteins from BA.2, BA.5, BA.2.75, BF.7, and XBB.1 to their receptor ACE2. In addition, they resolved high-resolution cryoEM structures of the S proteins from BA.2.75, BF.7, and XBB.1 bound to their receptor PD in order to conduct further research.
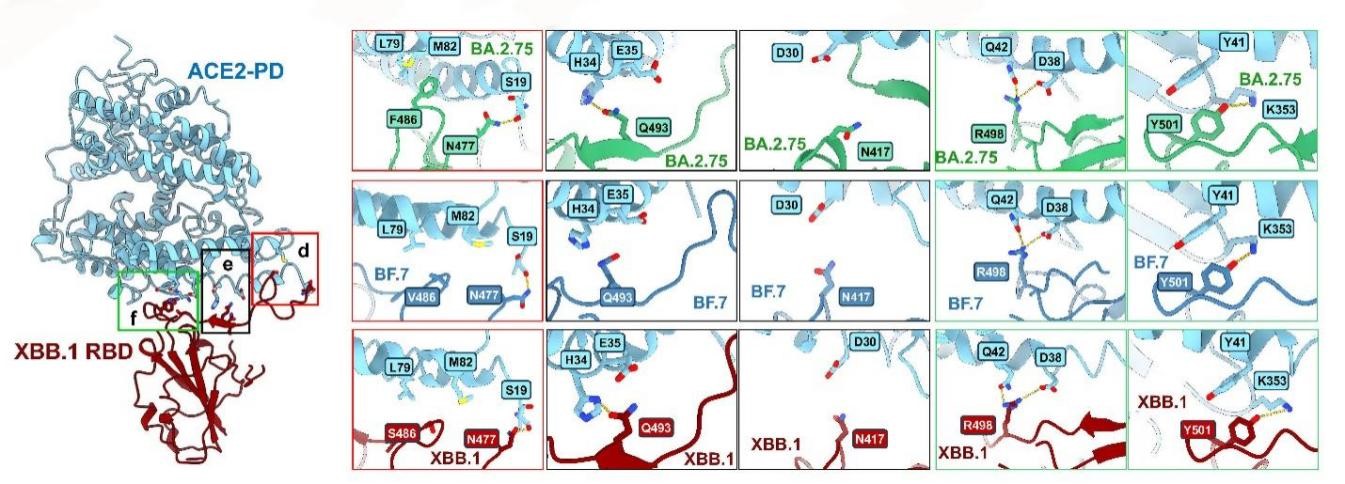
Figure 1. Interface analysis of the interaction between Omicron BA.2 derived mutant strains BA.2.75, BF.7, XBB.1 and receptor ACE2
To investigate the potential effects of N343 glycans on antibody neutralization, the researchers studied the N-linked glycosylation profiles of RBD and trimeric S proteins from SARS-CoV-2 WT, BA.5, and XBB.1 variants. Findings from the above series of studies suggest that mutations in the RBD region may affect N343 glycan composition by altering the glycan modification mechanism.
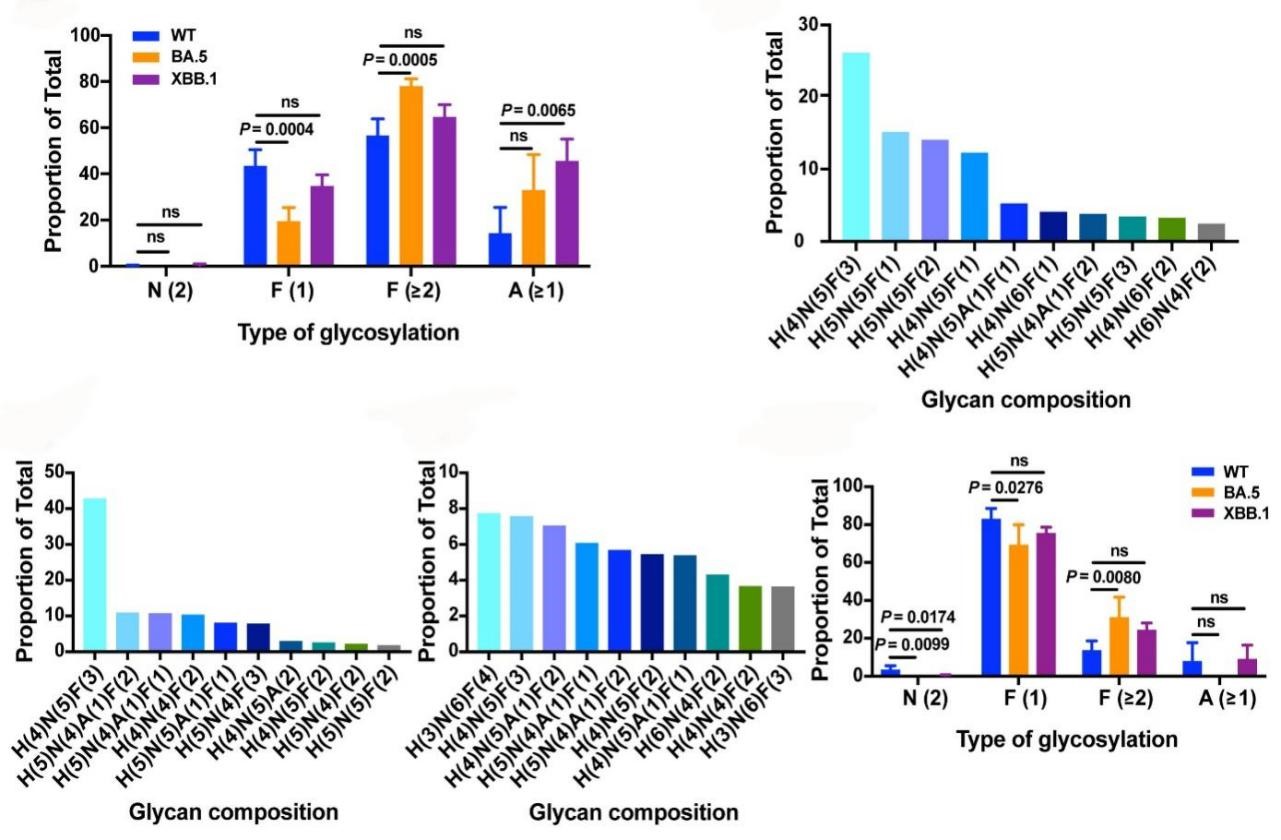
Figure 2. Glycosylation profiles of the Omicron BA.2-derived variants BA.2.75, BF.7, and XBB.1 at N343
In conclusion, Omicron variants carry a large number of mutations, resulting in their high transmissibility and immune evasion capabilities. Despite carrying these mutations, BA.2.75, BF.7, and XBB.1 still use ACE2 as the receptor, indicating that monoclonal antibodies mimicking ACE2 may have the broadest spectrum of neutralizing potency. Moreover, the mass spectrometry analysis of glycosylation modifications shows that the intact S protein expressed in vitro and the glycosylation modification expressed by the RBD region alone are different. This result provides a theoretical reference for vaccine development.
Yaning Li, a Ph.D. student from Tsinghua University, Chang Ren, a master’s student from SUSTech, and Yaping Shen and Yuanyuan Zhang, Ph.D. students from Westlake University, are co-first authors of this paper. Assistant Professor Renhong Yan and Associate Professor Liwei Cao are the co-corresponding authors.
The study was supported by the National Natural Science Foundation of China (NSFC), Science, Technology and Innovation Commission of Shenzhen Municipality, Cryo-Electron Microscopy Platform of Westlake University, and the Cryo-Electron Microscopy Center of SUSTech. Mass spectrometry analysis were provided by the Core Research Facilities of SUSTech.
Paper link: https://www.nature.com/articles/s41421-023-00607-2
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By