Argonaute proteins (Agos) play a crucial role in RNA silencing in eukaryotes by using small RNAs or DNAs as guides. In prokaryotes, particularly in the SPARTA immune system, Agos trigger cell death in response to invading plasmids, but its molecular mechanisms remain unknown and requires further investigation.
Associate Professor Ning Jia’s research group from the School of Medicine at the Southern University of Science and Technology (SUSTech) has recently published their study that identifies the molecular basis for target ssDNA-mediated SPARTA activation, offering insights that could facilitate the development of SPARTA-based biotechnological tools.
Their research work, entitled “Target ssDNA activates the NADase activity of the prokaryotic SPARTA immune system”, has been published in Nature Chemical Biology.
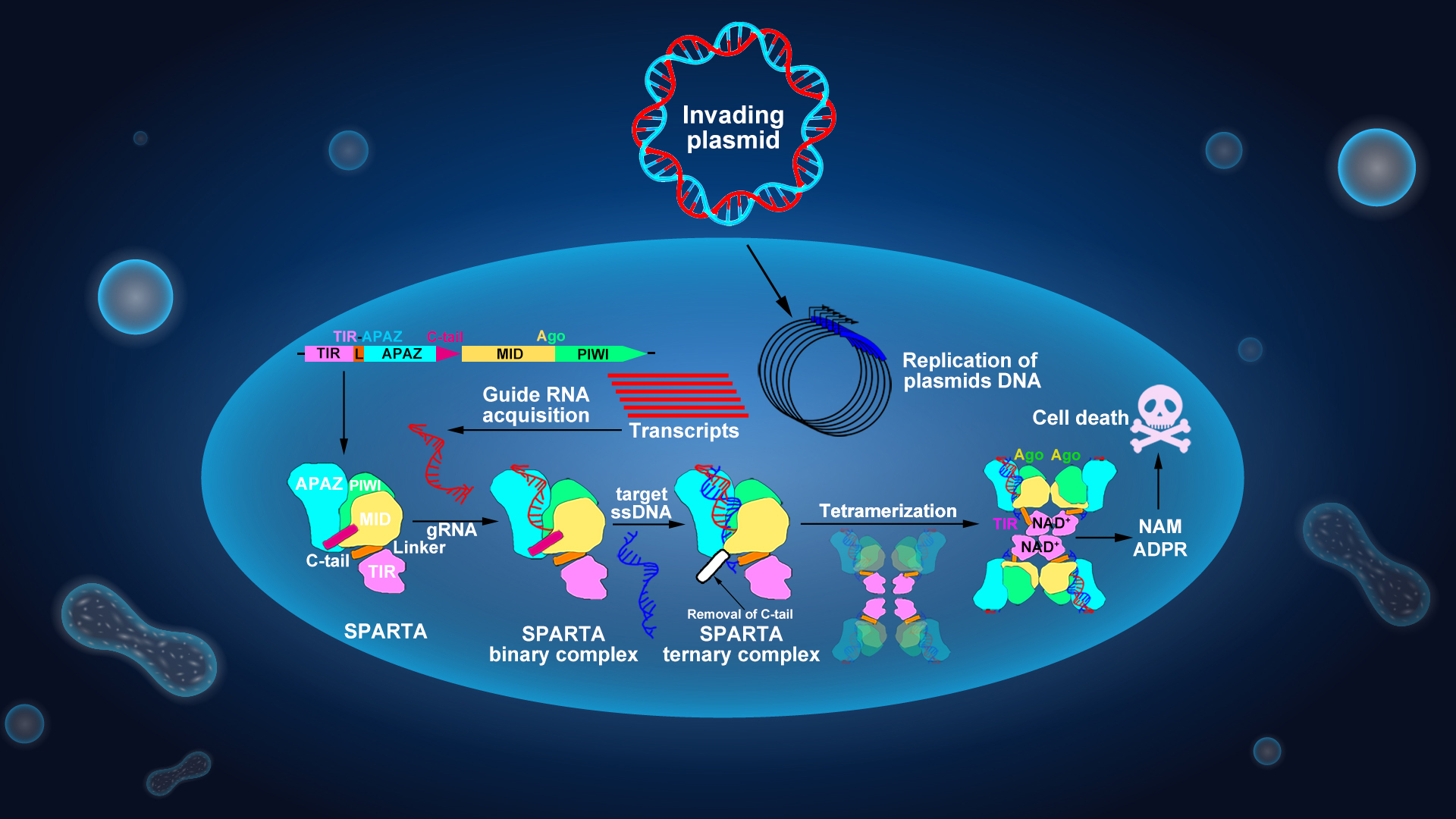
The group focused on elucidating the mechanism of the prokaryotic immune systems against the invading phages and plasmids, and has published a series of related papers (Molecular Cell, 2023, 2019a, 2019b, 2019c; Nature Communications, 2022; Science, 2020; Cell Research, 2020, 2022; Nature Reviews Molecular Cell Biology, 2021). The short prokaryotic Ago/TIR-APAZ (SPARTA) system is such a prokaryotic short pAgo immune system that contains two components: a short pAgo protein and a TIR-APAZ protein containing a TIR domain fused to the APAZ domain. Upon detection of invading DNA, the SPARTA complex oligomerizes and degrades essential NAD(P)+ to noncyclic ADPR(P) and NAM, leading to cell death. Still, the mechanisms of their action remain unclear.
To explore the mechanism of how this system defends against plasmid invasion, the researchers first confirmed that the two proteins that constitute the SPARTA system, TIR-APAZ and pAgo, can form a stable heterodimeric complex through biochemical experiments. This complex further recognizes the invading target ssDNA through the base-pairing with the guide RNA, binding with the nucleic acids duplex, triggering the formation of a tetramer complex, which unleashes the NADase activity. Using cryogenic electron microscopy (cryoEM), they determined the structures of the inactive monomeric SPARTA complex and the active tetrameric SPARTA complex bound to ssDNA targets. Supported by biochemical data, the structural analyses reveal mechanisms by which a small gRNA facilitates recognition of the complementary ssDNA target by the SPARTA complex, and ssDNA target binding triggers SPARTA tetramerization and subsequent NADase activation. These results provide a mechanistic understanding of prokaryotic SPARTA-mediated immunity against invading plasmids.
CryoEM analysis of the oligomeric SPARTA yielded reconstruction of the tetrameric SPARTAgRNA-ssDNA complex. The overall architecture of the tetrameric SPARTA complex is arranged in a “dimers of dimers” X-shape, whereby protomers 1 and 2, as well as protomers 3 and 4, formed basic dimers through short pAgo-mediated interactions. Two such dimers were then arranged into a tetrameric state via different interactions between the two TIR-interfacing dimers.
Compared to the inactive monomeric SPARTA complex, protomers 2 and 4 adopt an identical conformation, while the Linker and TIR domains in protomers 1 and 3 undergo 90° and 180° rotations, respectively, enabling the tetramerization of the TIR domains. Rearrangement of the TIR domains in protomers 1 and 3 enables the four TIR domains to form an asymmetric, two-stranded, head-to-tail, parallel arrangement at the center of the complex, while the remaining domains, along with the gRNA-ssDNA target duplex, were positioned outside the TIR complex (Figure 1).
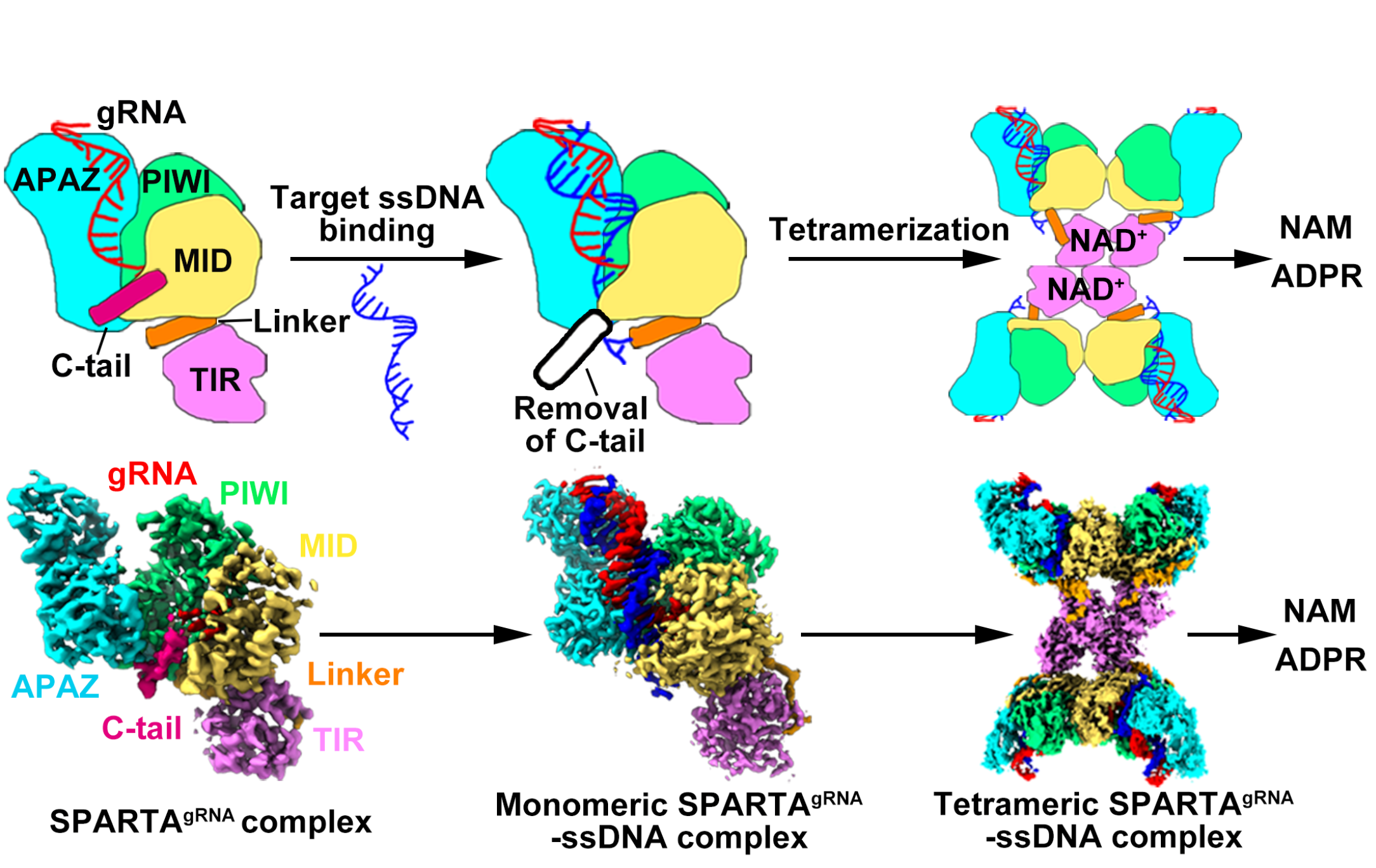
Figure 1. The assembly and activation of prokaryotic SPARTA immune system
In addition, the researchers further found that the SPARTA system harbors a highly conserved α-helix (C-tail) at its carboxyl terminus. This acidic α-helix is localized within the basic channel where the SPARTA system binds nucleic acid duplexes, and plays an auto-inhibitory role. Structural analysis of the SPARTAgRNA binary complex revealed that the C-tail undergoes a slight movement when the guide RNA strand is binding to the SPARTA system. Then, the small RNA guides SPARTA to recognize its complementary ssDNA target, leading to the removal of the inhibitory C-tail and the following SPARTA tetramerization. The hydrolysis of NAD+ into ADPR and NAM by SPARTA triggers the suicide of the invaded cell, thereby protecting the rest of the bacterial population.
In conclusion, this study elucidates the molecular mechanisms of prokaryotic SPARTA immune systems against invading plasmid DNA. The short pAgo and TIR-APAZ proteins form a stable SPARTA effector complex. The short pAgo serves as a sensor to bind a small RNA guide, which recognizes its complementary ssDNA target. The binding of target ssDNA by the short pAgo triggers SPARTA tetramerization and rearrangement of its TIR domains, leading to the formation of a catalytic pocket where NAD+ hydrolysis subsequently occurs (Figure 2). These findings deepen our understanding of prokaryotic SPARTA defense systems and provide a molecular basis for the development of SPARTA-based biotechnological tools.
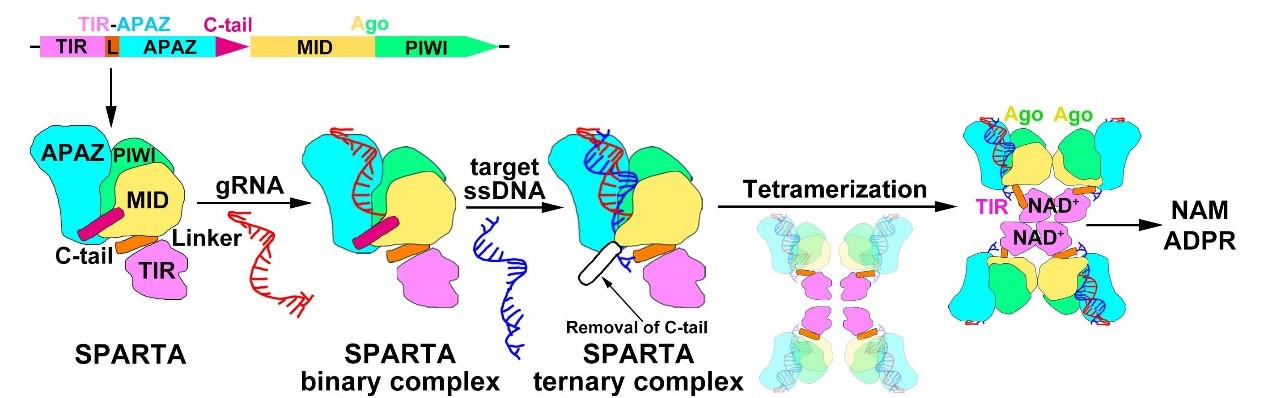
Figure 2. Proposed mechanistic model of prokaryotic SPARTA complex activation in response to invading plasmid DNA
Jun-Tao Zhang and Xin-Yang Wei are the co-first authors of this paper. Associate Professor Ning Jia from the Department of Biochemistry, School of Medicine at SUSTech is the corresponding author, and SUSTech is the first affiliated unit.
This work was supported by the National Natural Science Foundation of China (NSFC), Shenzhen and Guangdong Natural Science Foundation, Shenzhen Peacock Program, and Guangdong Provincial Science and Technology Innovation Council Grant. The researchers also acknowledge the work of the Cryo-EM Center at SUSTech for providing assistance in data collection and data processing.
Paper link: https://www.nature.com/articles/s41589-023-01479-Z
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo BySchool of Medicine