Over the past decade, clustered regularly interspaced short palindromic repeats-Cas (CRISPR-Cas) systems have been rapidly developed for gene editing in eukaryotes, contributing significantly to biomedical research, gene therapy, and plant and animal breeding.
Beyond the well-established type II Cas9 system, other Cas proteins, such as the type V-I Cas12i family, have also been identified and characterized. Compared to Cas9 and Cas12a, Cas12i proteins have a relatively smaller size, can process pre-crRNA into mature crRNA guides, and recognize AT-rich PAM sequences. However, naturally, Cas12i proteins often exhibit low efficiency and narrow targeting ranges, limiting their applications.
Professor Jian-Kang Zhu’s research group from the Institute of Advanced Biotechnology and the School of Medicine at the Southern University of Science and Technology (SUSTech) has recently reported a novel method based on AI-guided engineering of a Cas12i nuclease enables robust gene editing in animal and plant.
Their study, entitled “An engineered Cas12i nuclease that is an efficient genome editing tool in animals and plants”, has been published in The Innovation, an interdisciplinary journal covering scientific disciplines such as physics, chemistry, materials, nanotechnology, biology, translational medicine, geoscience, and engineering.
The researchers first predicted the 3D structure of Cas12i3 using AlphaFold2, and compared it with structures of related Cas12i1/i2 proteins to predict key amino acid residues for its interaction with nucleic acids. They then substitute them with the positively charged arginine (R) and build a library of 150 single-point mutation variants. They screened 26 Cas12i3 variants with activity increased by at least 1.5-fold using a fluorescent reporter system.
Through further combining point mutations in the Cas12i3 REC domain and WED domain, the researchers obtained Cas-SF01 (S7R/D233R/D267R/N369R/S433R) with activity increased by about 4-fold than wildtype Cas12i3 in mammalian cells. At multiple endogenous target sites, Cas-SF01 exhibited an average indel rate of approximately 80%, surpassing the indel rate of hfCas12Max (~70%), AsCas12a Ultra (~60%), and SpCas9 (~60%). Further characterization found that Cas-SF01 effectively recognizes canonical NTTN PAMs and non-canonical NATN, TTVN PAMs. They also engineered a high-fidelity version, Cas-SF01HiFi, by introducing a single mutation, D876R, which reduces off-target effects while maintaining high on-target activity.
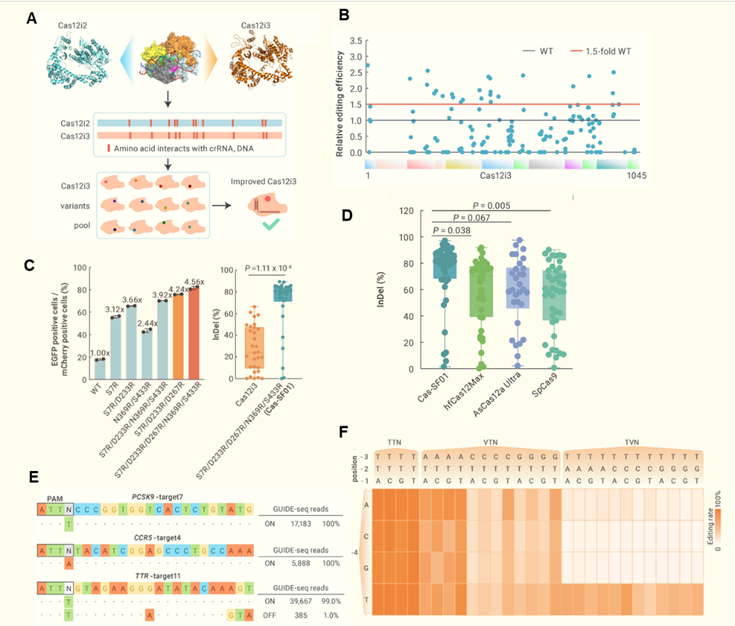
Figure 1. Engineering high-performance of Cas-SF01
Professor Zhu’s group evaluated the in vivo gene editing capability of Cas-SF01 by delivering it along with guide RNAs using lipid nanoparticles (LNPs) to mouse liver. Cas-SF01 achieved editing efficiencies at the target Ttr locus comparable to the commonly used SpCas9 and previously reported hfCas12Max systems. This demonstrates the robustness of Cas-SF01 for in vivo therapeutic gene editing applications. They next explored the use of Cas-SF01 for plant gene editing by generating stable transgenic lines in rice and hairy root systems in pepper and soybean. In rice, Cas-SF01 enabled highly efficient editing, with average indel mutation rates reaching ~75% across multiple targets. Strikingly, while no editing was detected with wildtype Cas12i3 in pepper or soybean hairy roots, Cas-SF01 achieved editing efficiencies of ~50% in both plant species. Together, their rational engineering strategy empowered a previously low-activity Cas12i3 nuclease into a versatile, high-efficiency gene editing platform applicable to both animal and plant systems. Cas-SF01 and its high-fidelity variant Cas-SF01HiFi can serve as robust tools for therapeutic gene and cell editing as well as accelerating molecular breeding efforts in diverse organisms.
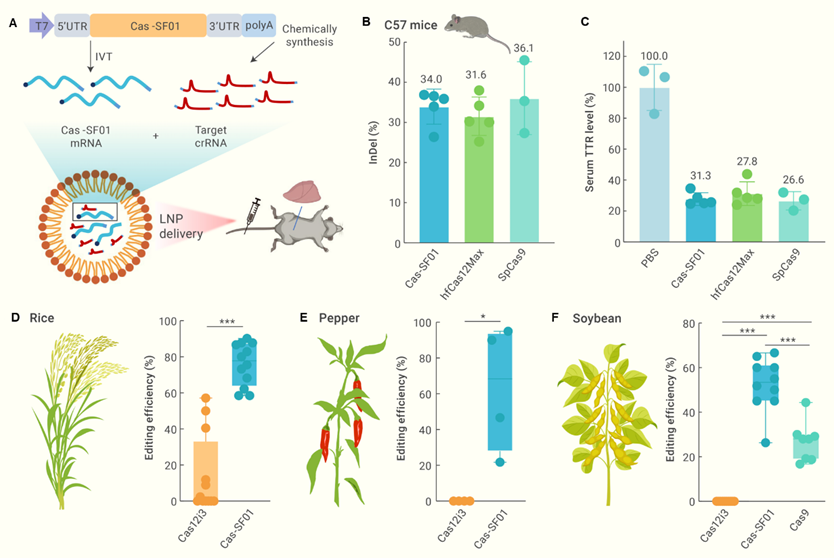
Figure 2. Highly efficient editing in animals and plants
Dr. Zhiqiang Duan and Yafeng Liang, a postdoctoral fellow and visiting scholar in Professor Jian-Kang Zhu’s group, are the co-first authors of this paper. Professor Jian-Kang Zhu is the corresponding author. Dr. Jialei Sun and Dr. Yechun Hong from the Institute of Advanced Biotechnology and School of Medicine, and Assistant Professor Ruiheng Tian from the School of Medicine at SUSTech, also provided important experimental support in this study.
This work was supported by the National Natural Science Foundation of China (NSFC) and Bellagen Biotechnology Co. Ltd., Jinan.
Paper link: https://www.cell.com/the-innovation/fulltext/S2666-6758(24)00002-X
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo ByInstitute of Advanced Biotechnology