Neutralizing antibodies are crucial for preventing and treating COVID-19. However, numerous Omicron subvariants continue to emerge and impair the efficacy of existing neutralizing antibodies. The speed of antibody development lags behind the mutation rate of new SARS-CoV-2 variants.
Most commercially available neutralizing antibodies developed in the early stages have been largely eliminated even before the clinical trials were completed. The search for ultra-conservative antigenic epitopes, the screening of specific neutralizing antibodies, and the modification of different targeted antibody combinations are crucial directions to reduce or avoid immune escape. Additionally, the discovery and modification of conservative epitopes also contribute to the development of broad-spectrum SARS-CoV-2 vaccines.

In a previous study, scientists have constructed various specific and broad-spectrum neutralizing antibodies, including the development of a dual-specific antibody against SARS-CoV-2 variants, BA7208/7125 (Cell Discovery, 2023). To address the challenge of the continuous emergence of mutations in the new Omicron variants, Professor Zheng Liu from the Cryo-Electron Microscopy Center at the Southern University of Science and Technology (SUSTech), in collaboration with the National Clinical Research Center for Respiratory Disease at the First Affiliated Hospital of Guangzhou Medical University, the Antibody Research and Development Center at Shandong Boan Biotechnology Co., Ltd., and the Division of Monoclonal Antibodies at the National Institutes for Food and Drug Control (NIFDC) has analysed the conservative epitopes in the spike protein of the SARS-CoV-2 variants, developing a high-efficient and broad-spectrum neutralizing antibody.
The team successfully identified a highly conserved neutralizing epitope targeted by an antibody BA7535, which demonstrates high neutralization potency against not only previous variants, such as Alpha, Beta, Gamma, Delta and Omicron BA.1-BA.5, but also more recently emerged Omicron subvariants, including BF.7, CH.1.1, XBB.1, XBB.1.5, XBB.1.9.1, and EG.5. Structural analysis of the Omicron spike protein with BA7535 using cryo-EM indicates that the antibody recognizes a highly conserved cryptic receptor-binding domain (RBD) epitope, avoiding most of the mutational hot spots in the RBD. Furthermore, structural simulation based on the interaction of BA7535 /RBD complexes dissects the broadly neutralizing effect of BA7535 against latest variants. Therapeutic and prophylactic treatment with BA7535 alone or in combination with BA7208 protected mice from the circulating Omicron BA.5 and XBB.1 variant infection, suggesting the highly conserved neutralizing epitope serves as a potential target for developing highly potent therapeutic antibodies and vaccines.
The results, entitled “Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants”, have been published in the international journal Nature Communications.
In this study, scientists utilized immunizing human antibody transgenic mice combined with phage display technique and neutralization experiments. Large-scale screening from the immunization library identified multiple broad-spectrum candidate antibodies (Figure 1). Their characteristics and neutralizing activities were determined through IC50, affinity KD, and ADCC and ADCP assays. Among the antibodies, the BA7535 showed superior neutralization efficiency compared to other commercially available antibodies (Figure 2).
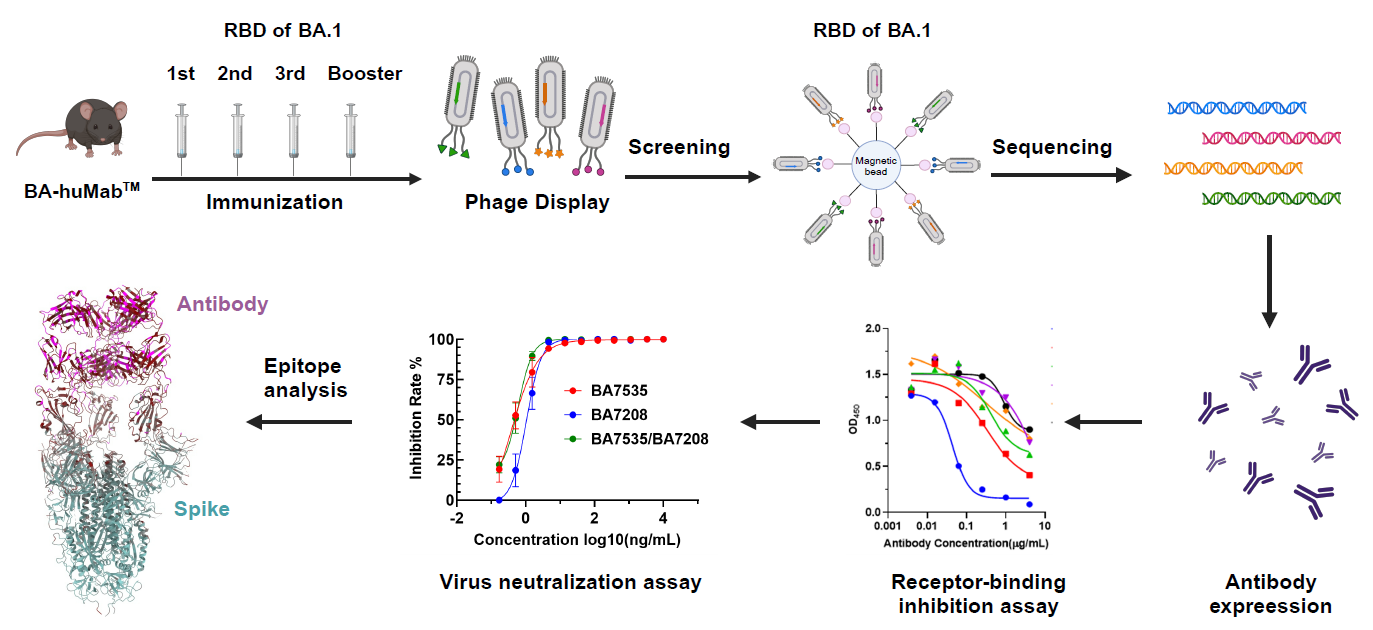
Figure 1. Broad-spectrum antibody screening strategy
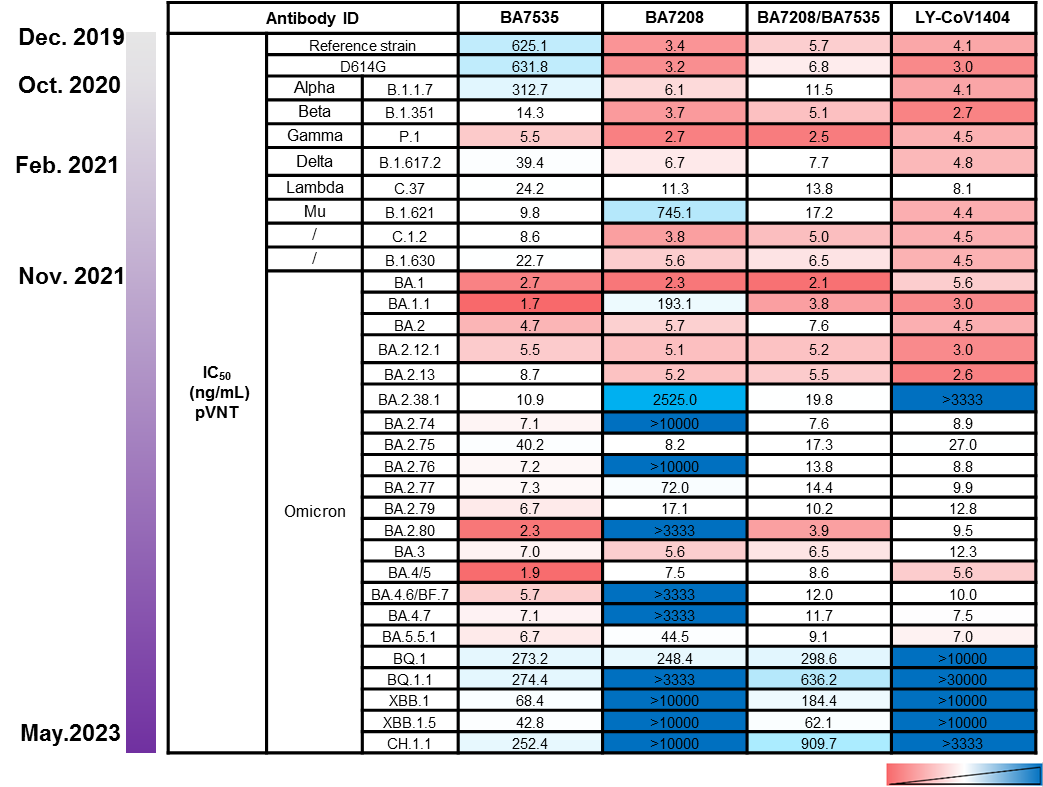
Figure 2. Comparison of the activity of highly efficient broad-spectrum antibodies. BA7535 with other antibodies
The cryo-EM structure of the antigen-antibody complex revealed that the BA7535 targeted a relatively conservative epitope present in almost all Omicron variants, involving seven key amino acid residues, including T415, D420, Y421, A475, N487, Y489, and R493. Multiple salt bridges and hydrogen bonds were formed during this binding. The epitope was located at the top of the RBD, partially overlapping with the ACE2 epitope, elucidating that the BA7535-Fab, by binding to the RBD of the Spike, occupies the binding site of the RBD to ACE2, which in turn directly blocks the binding of ACE2 to the RBD and thus blocking the virus invasion into host cells (Figure 3).
Structural simulation and analysis experiments predicted that the BA7535 antibody had no obvious immune escape against emerging mutant strains of the Omicron variants. The epitope of BA7535 showed a very low frequency in the virus mutation monitoring analysis. Comparative analysis of epitopes of BA7535 and 221 published RBD-specific antibodies indicated that most of the previously reported neutralizing antibodies target one or two major antigenic sites. Single or double mutations of concern at key positions always lead to immune escape, while the epitope residues and buried surface area (BSA) for each epitope residue of BA7535 are more dispersive and appear more resistant to most mutants. A comprehensive analysis of the impact of 42 sub-lineages of Omicron indicated that BA7535 has high-efficient binding and neutralizing potential against all 42 sub-lineages (Figure 4).
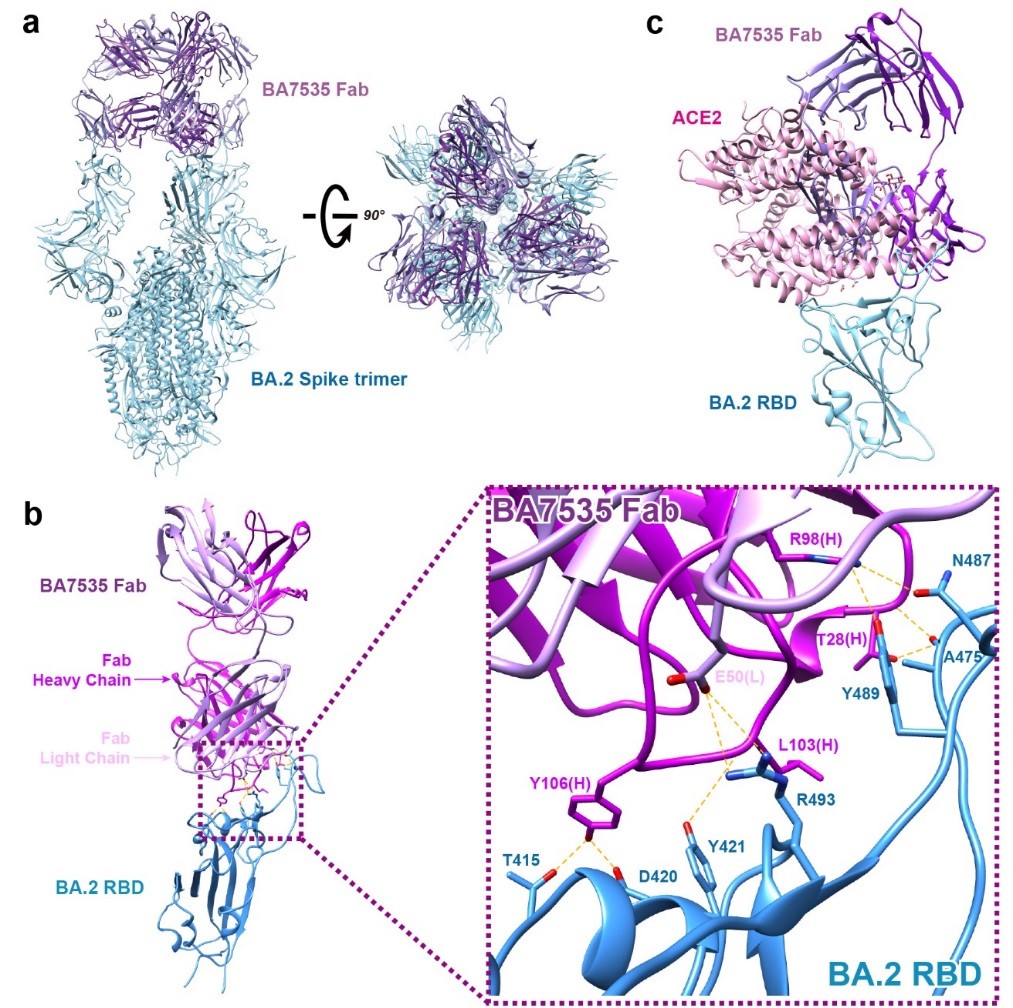
Figure 3. Cryo-electron microscopy of Omicron BA.2 Spike protein with BA7535
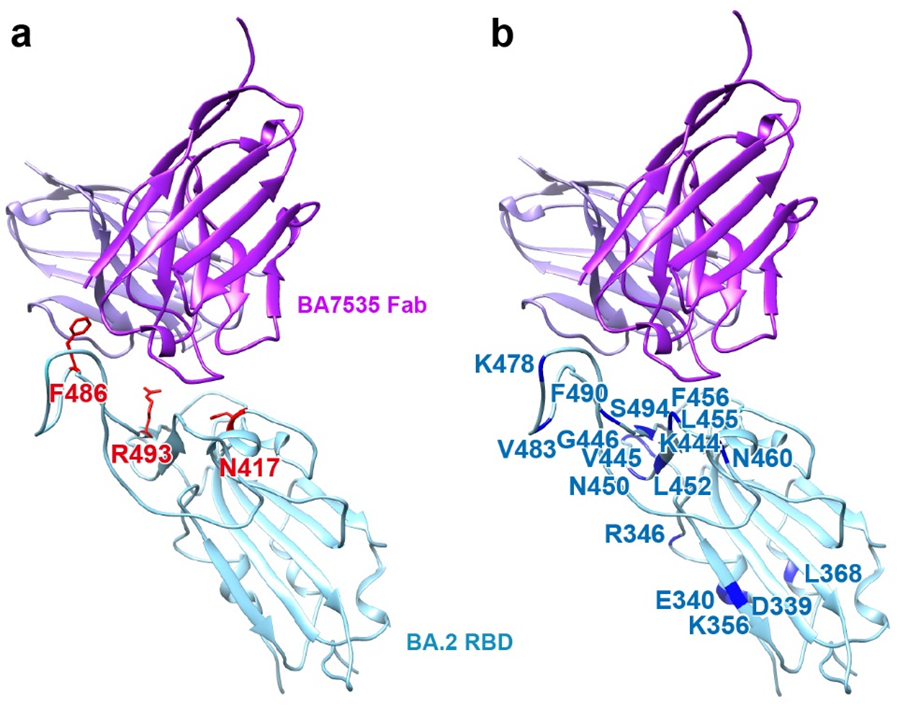
Figure 4. BA7535 binding site avoids most mutations from Omicron sub-lineages
In vivo protection experiments demonstrated that the BA7535 antibody could effectively prevent and treat infections in mice, including Omicron BA.5 and XBB.1. Moreover, diverse administration routes, including intraperitoneal injection (i.p), intranasal (i.n), and aerosol inhalation (a.i), all provided efficient protection (Figure 5).
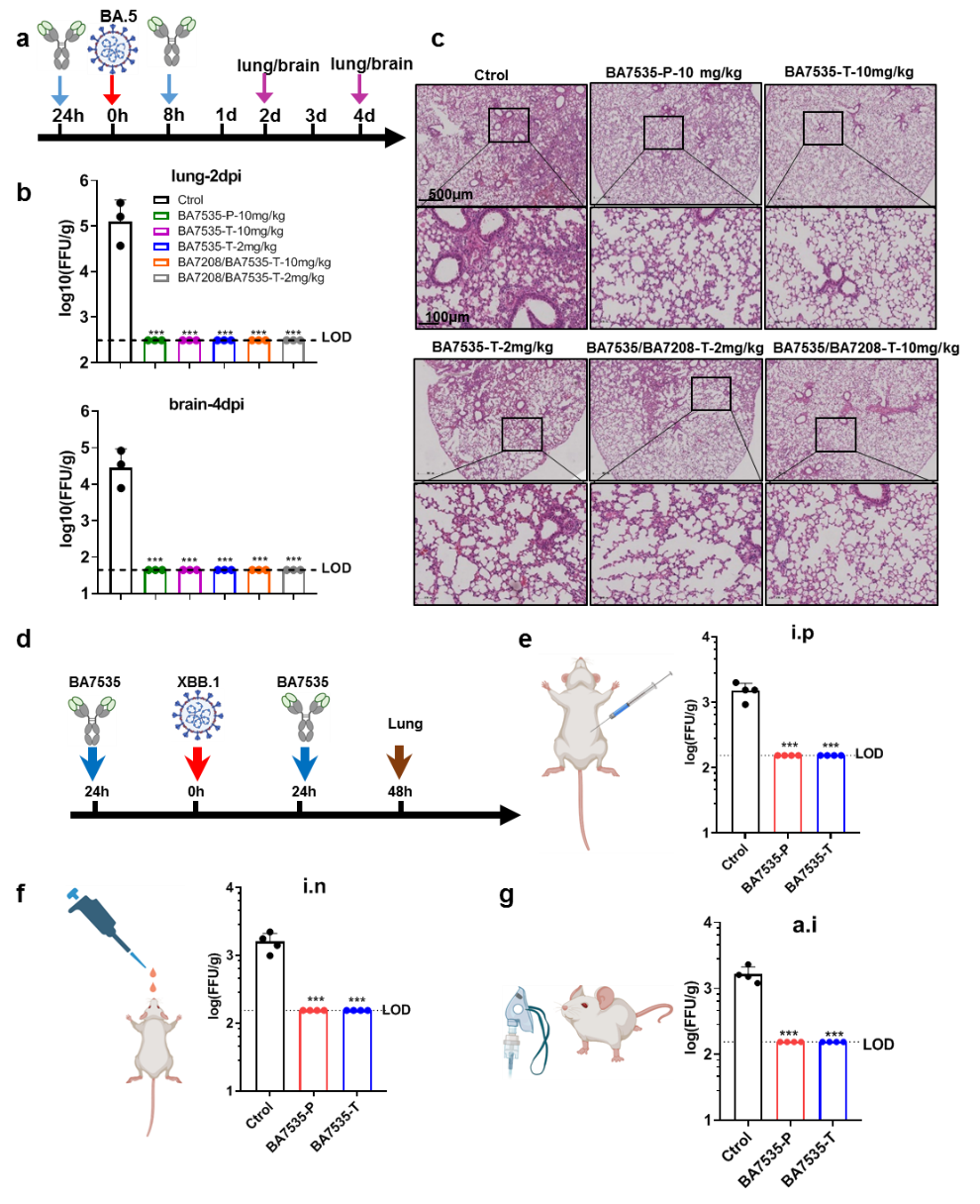
Figure 5. BA7535 protected mice from Omicron BA.5 and XBB.1 variant infection
Escape mutant screening experiments revealed that both BA7535 and BA7208 (a previously developed broad-spectrum neutralizing antibody, Cell Discovery, 2023) showed weak escape. Surprisingly, no dominant escape mutants were observed in the spike protein, indicating that the BA7535/BA7208 cocktail leads to less selective pressure compared to single-antibody treatment (Figure 6).
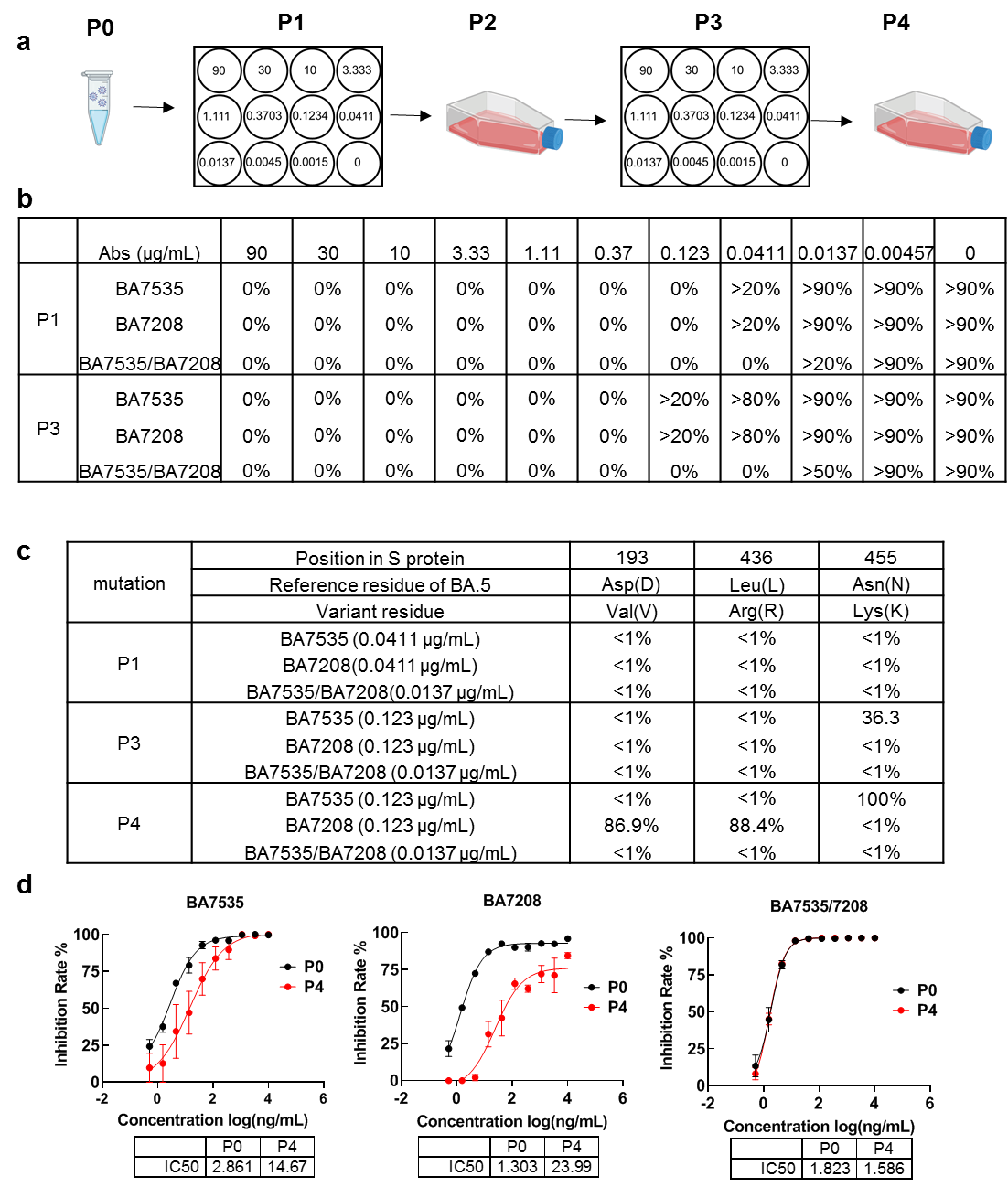
Figure 6. Escape mutant screening under the pressure of antibodies
Prof. Yanqun Wang from the First Affiliated Hospital of Guangzhou Medical University, Research Assistant An Yan from the Cryo-EM Center at SUSTech, Dr. Deyong Song from Boan Biotechnology Co., Ltd., and Dr. Maoqin Duan from NIFDC are the co-first authors of the paper. Prof. Jincun Zhao from Guangzhou Medical University/Guangzhou National Laboratory, Prof. Zheng Liu from the Cryo-EM Center at SUSTech, Dr. Changlin Dou from Boan Biotechnology Co., Ltd., and Dr. Lan Wang from NIFDC are the co-corresponding authors.
The cryo-EM data collection, processing, structure analysis, and interpretation were performed at the Cryo-EM Center at SUSTech, with the assistance of Senior Engineers, including Dr. Yuanzhu Gao and Dr. Xiaomin Ma, and Research Assistant Professor Ruxi Qi.
This project was supported by the National Key R&D Program of China, National Natural Science Foundation of China (NSFC), and the Guangdong Basic and Applied Research Projects.
Paper link: https://www.nature.com/articles/s41467-024-45050-3
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo ByCryo-Electron Microscopy Center