β-Coronaviruses are a group of positive single-strand RNA viruses which include the pathogenic strains severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), SARS-CoV, murine hepatitis virus (MHV), and the Middle East respiratory syndrome coronavirus (MERS-CoV), etc. After entering the cell, the virus utilizes the host’s translation system to rapidly synthesize a series of viral proteins, including multiple nonstructural proteins (Nsp). Among them, Nsp3 and Nsp4 induce the reshaping of the host cell’s membrane system, forming replication organelles (RO), mainly structured as double-membrane vesicles (DMV).
DMVs provide a secure environment for viral RNA synthesis and storage, facilitating efficient viral genome replication and translation. Previous studies showed that most DMVs are tightly packed into clusters during virus infection. The molecular mechanism underlying the spatial confinement of DMVs remains unknown.
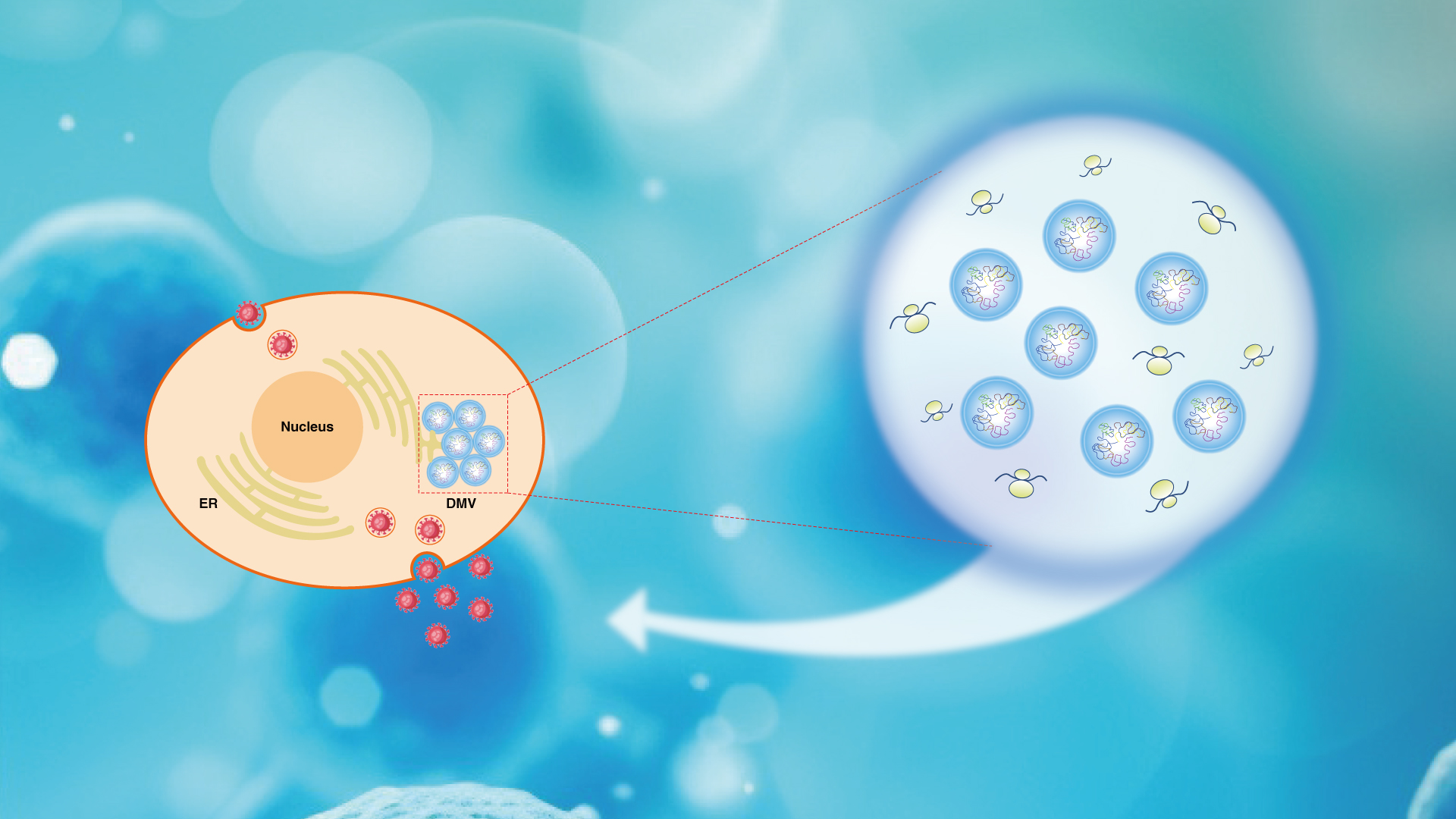
Associate Professor Yan Zhao’s research group from the School of Life Sciences, in collaboration with Professor Zheng Zhang’s group from the Institute for Hepatology, National Clinical Research Center for Infectious Disease, Shenzhen Third People’s Hospital at the Southern University of Science and Technology (SUSTech), have revealed that SARS-CoV-2 exploits host FXR (fragile X-related) family proteins, which undergo liquid-liquid phase separation (LLPS) for gathering DMVs and recruiting translation machinery simultaneously to facilitate viral protein synthesis and achieve efficient viral replication.
Their research results were published in the Journal of Cell Biology, entitled “LLPS of FXR proteins drives replication organelle clustering for β-coronaviral proliferation”.
The FXR family proteins, including FXR1, FXR2, and FMR1/FMRP, play important roles in RNA metabolism, neuronal homeostasis, muscle development, and cancer occurrence. A recent study reported that FXR1 forms phase-separated liquid droplets, recruiting translation machinery to promote the translation of sperm-specific mRNAs crucial for late-stage sperm development. Here, the researchers discovered that FXR1/2 and FMR1 are recruited to DMV sites by binding to Nsp3 on the outer membrane of DMVs, a process mediated by the UBL1 and HVR domains of Nsp3 and the NIR (Nsp3 interacting region) segment of FXR1. In control cells, DMVs exhibit clustered distribution in the cytoplasm, while upon triple knockdown of FXR1/2 and FMR1 (siFXRs), DMVs show a dispersed pattern (as shown in Figure 1). These results indicate the essential function of the FXRs in clustering DMVs.

Figure 1. Exogenous expression of Nsp3 and Nsp4 induces the formation of DMV-like structures, with DMVs exhibiting clustered distribution in control cells, while upon siFXRs, DMVs disperse throughout the cytoplasm
In vitro experiments demonstrate that purified Nsp3 alone cannot undergo liquid-liquid phase separation, but it can enter droplets formed by FXR1 protein. FXR1 droplets concentrate Nsp3-decorated liposomes. Further investigation reveals that FXR1 droplets, both in vitro and in vivo, recruit viral RNA and ribosomes to promote local RNA translation. During SARS-CoV-2 infection, signals of FXR proteins, translation initiation factors, and ribosomal proteins are detected at DMV sites. Compared to control cells, siFXRs cells exhibit significantly reduced capacity for viral protein synthesis, leading to pronounced inhibition of viral replication (Figure 2).
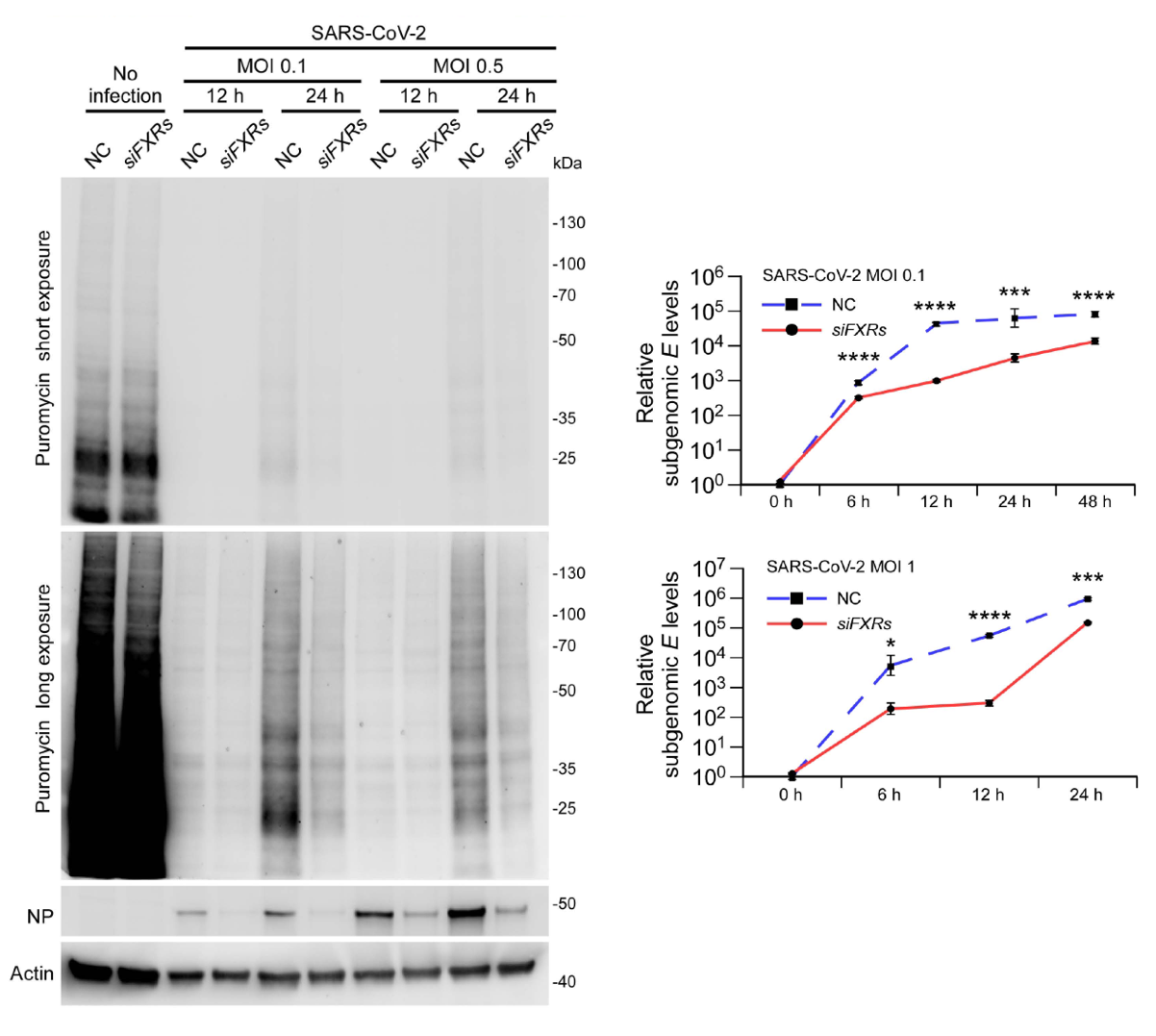
Figure 2. Viral protein synthesis and subgenomic E RNA levels are greatly reduced in siFXRs cells, compared to control cells after SARS-CoV-2 infection
In conclusion, the study reveals a novel model in which LLPS of FXRs mediates DMV compartmentalization for the assembly of viral replication centers and activation of viral RNA translation (Figure 3). This advance uncovers a potential therapeutic target for treating infectious diseases caused by SARS-CoV-2 or other viruses using similar mechanisms.
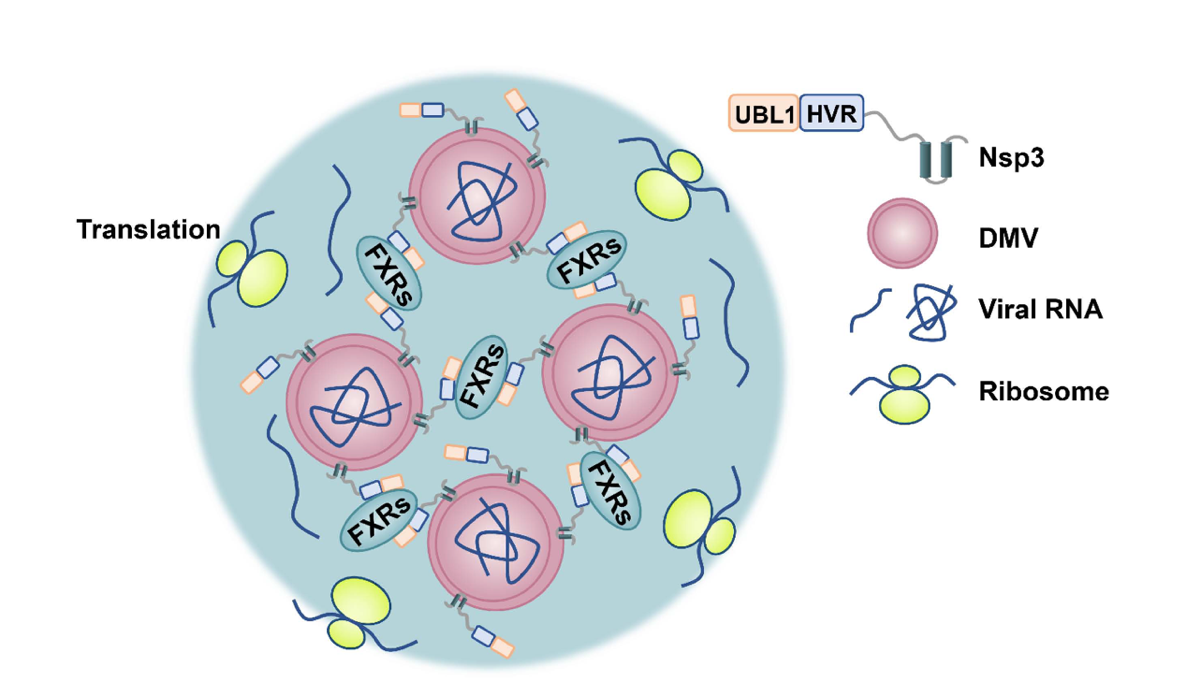
Figure 3. LLPS of FXRs drives clustering of DMVs and recruits translation machinery
Graduate students Meng Li, Yali Hou, and Zhenni Yang, along with postdoctoral fellow Yuzheng Zhou, are the co-first authors of this paper. Associate Professor Yan Zhao and Professor Zheng Zhang at SUSTech are the corresponding authors. Assistant Professor Fuxing Zeng, Research Associate Professor Xiaotian Liu, and graduate students Tao Jian and Qianxi Yu at SUSTech are the co-authors. Dr. Hongyu Zhao from the Institute of Biophysics of the Chinese Academy of Sciences and Dr. Lin Lin from the SUSTech Core Research Facilities provided technical support.
This work was supported by the National Key Research and Development Program, National Natural Science Foundation of China (NSFC), Shenzhen Science and Technology Program, and the Shenzhen–Hong Kong Institute of Brain Science–Shenzhen Fundamental Research Institutions.
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo BySchool of Life Sciences