Understanding DNA methylation and its transgenerational inheritance is essential for grasping how epigenetic changes influence genetic stability and adaptation. DNA methylation regulates gene expression and maintains genome integrity, but disruptions in demethylation can lead to harmful methylation patterns. Studying these mechanisms in Arabidopsis offers insights into epigenetic inheritance, crucial for enhancing crop resilience, improving agricultural productivity, and ensuring food security.
Further research could reveal parallels in other organisms, including humans, advancing knowledge in evolutionary biology, medicine, and developmental biology. Thus, exploring and investigating active DNA demethylation and its effects on epigenetic inheritance remains critical for both fundamental and applied sciences.
Professor Jian-Kang Zhu’s research group from the Institute of Advanced Biotechnology at the Southern University of Science and Technology (SUSTech) has recently published a study on the model organism Arabidopsis. The study found that a mutation in the DNA demethylase ROS1 led to a generational increase in DNA methylation at the genomic level. Their findings reveal the important monitoring role of ROS1 in preventing the transgenerational inheritance of randomly formed DNA methylation and maintaining the stability of epigenomes.
Their work, entitled “Transgenerational increases in DNA methylation in Arabidopsis plants defective in active DNA demethylation”, has been published in the Proceedings of the National Academy of Sciences (PNAS).
Under both normal and salt stress conditions, single-seed Arabidopsis descents were consecutively propagated for ten generations (G1-G10), and five independent lineages were created in each generation. This study systematically compared the transgenerational changes in DNA methylation under two genotypes (wild type and ros1 mutant) and two growth conditions (normal and salt stress).
The results showed that the average number of differentially methylated cytosines (DMCs) and the CG epimutation rate in the G10 ros1 mutant were above twice as high as those in the wild type, and the non-CG (CHG and CHH)-DMCs were also higher than those in the wild type. In the wild type, transgenerational DMCs with increased methylation accounted for approximately 50% of total transgenerational DMCs, while this proportion in the ros1 mutant was much higher.
These results indicated that active DNA demethylation controlled the transgenerational increases in DNA methylation, thereby maintaining the stability of epigenomes.
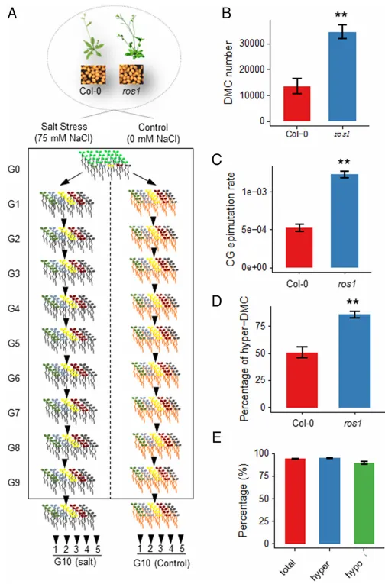
Figure 1. Experimental design and transgenerational DMC analysis
Interestingly, the transgenerational DMCs in the ros1 mutant were not the conventional ROS1 target regions. Differentially methylated regions (CG-DMRs) showed consistent transgenerational DNA methylation changes in the five ros1 mutant replicates. In contrast, the wild type showcased random changes. Consistent with the changing trend of transgenerational CG-DMCs, CG-DMRs in the ros1 mutant were mostly hyper-CG-DMRs, and the proportions of hyper-CG-DMRs of the wild type and the ros1 mutant in total CG-DMRs were 54% and 99%, respectively.
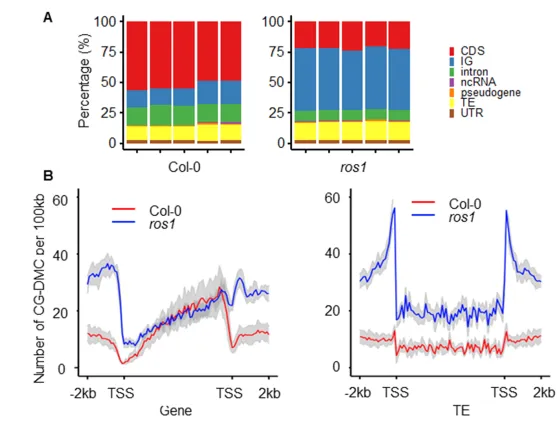
Figure 2. Characterization of the transgenerational CG-DMCs in the wild type and the ros1 mutant under normal growth conditions
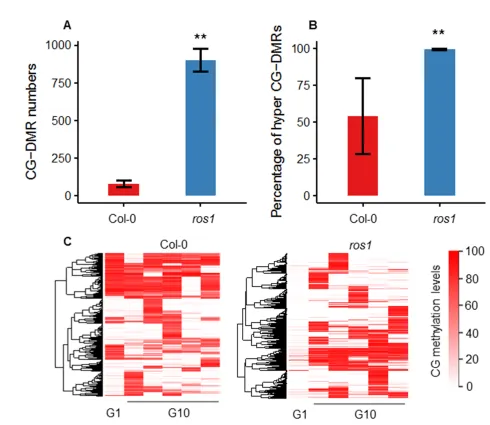
Figure 3. Analysis of the transgenerational CG-DMRs in the wild type and the ros1 mutant under normal growth conditions
The researchers also found that under salt stress conditions, the wild type and the ros1 mutant exhibited the characteristics of transgenerational CG-DMRs similar to those under normal conditions. This demonstrated the important role of active demethylation in maintaining the stability of transgenerational epigenomes under different conditions. Therefore, they propose that ROS1 is an important monitoring factor that prevents random DNA methylation transgenerational inheritance.
This finding provides clues for future research on human transgenerational inheritance, genomic DNA methylation evolution, and the identification of key factors in maintaining stable transgenerational inheritance of genomic DNA methylation. Additionally, this study will help us understand the evolution of genomic DNA methylation and how the environment affects offspring through epigenetic inheritance.
Dr. Kai Tang, a former doctoral student of Professor Jian-Kang Zhu, is the first author of the paper. Professors Jian-Kang Zhu and Zhaobo Lang from SUSTech are the corresponding authors. Other contributors to this work included Professor Xiaohong Zhu and Dr. Shaojun Xie (former team members of Jian-Kang Zhu’s lab).
Paper link: https://www.pnas.org/doi/10.1073/pnas.2320468121
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo ByInstitute of Advanced Biotechnology