Similar to the framework of living buildings, cells own the cytoskeleton system to provide structural support for functional activities, including to maintain cell shape and size and regulate cell movement and division. Actin, a key component of the cytoskeleton, facilitates the highly dynamic polymerization and depolymerization between its monomeric and filamentous forms in cells, which is vital for various cellular activities. However, besides actin itself, some regulatory proteins were discovered to modify actin dynamics.
The MICAL family was recently identified as a novel class of F-actin depolymerization factors with monooxygenase enzymatic activity. These factors facilitate actin cytoskeleton depolymerization by covalently oxidizing the two special methionine residues between actin subunits, with the assistance of its cofactors FAD and NADPH. Unlike the traditional actin regulatory factors, due to the enzymatic activity for covalent modification, MICAL proteins perform highly efficiently to locally regulate actin dynamics in cells, and loss-of-control of MICAL enzymatic activity would cause severe diseases.
Uncovering the regulation mechanism of MICAL proteins is essential for understanding actin cytoskeleton dynamics as well as cytoskeleton-involved disorders. However, due to the complexity and flexibility of MICAL proteins, understanding the activity regulation at an entire molecule scale is still challenged. In the past years, scientists have struggled to uncover how the activity of MICAL proteins are tightly controlled, expecting to further understand actin cytoskeleton dynamics in the complicated “building” of the cells.
A research team led by Associate Professor Zhiyi Wei from the Department of Neuroscience, School of Life Sciences at the Southern University of Science and Technology (SUSTech) has recently published a study revealing the three-dimensional cryo-EM structure of the full-length MICAL1 protein, a novel F-actin depolymerization regulator. Their work uncovers the molecular mechanisms of autoinhibition formation and its relief, which control MICAL1’s enzymatic oxidase activity in actin cytoskeleton dynamics.
Their paper, entitled “Autoinhibition and Relief Mechanisms for MICAL Monooxygenases in F-actin Disassembly”, has been published in Nature Communications.
The researchers first revealed the autoinhibition formation and relief mechanisms for MICAL family proteins in the regulation of enzymatic activity, contributing to a breakthrough in the field. After extensive efforts, they successfully expressed and purified a high-quality, full-length MICAL1 protein sample in vitro. It was detected as a monomeric form with relatively low enzymatic activity, indicating that the purified MICAL1 protein is autoinhibited in solution. By employing cryo-EM single particle analysis, they determined the three-dimensional structure of full-length MICAL1 protein, exhibiting an autoinhibitory conformation formed by multiple domains assembled together (Fig. 1).
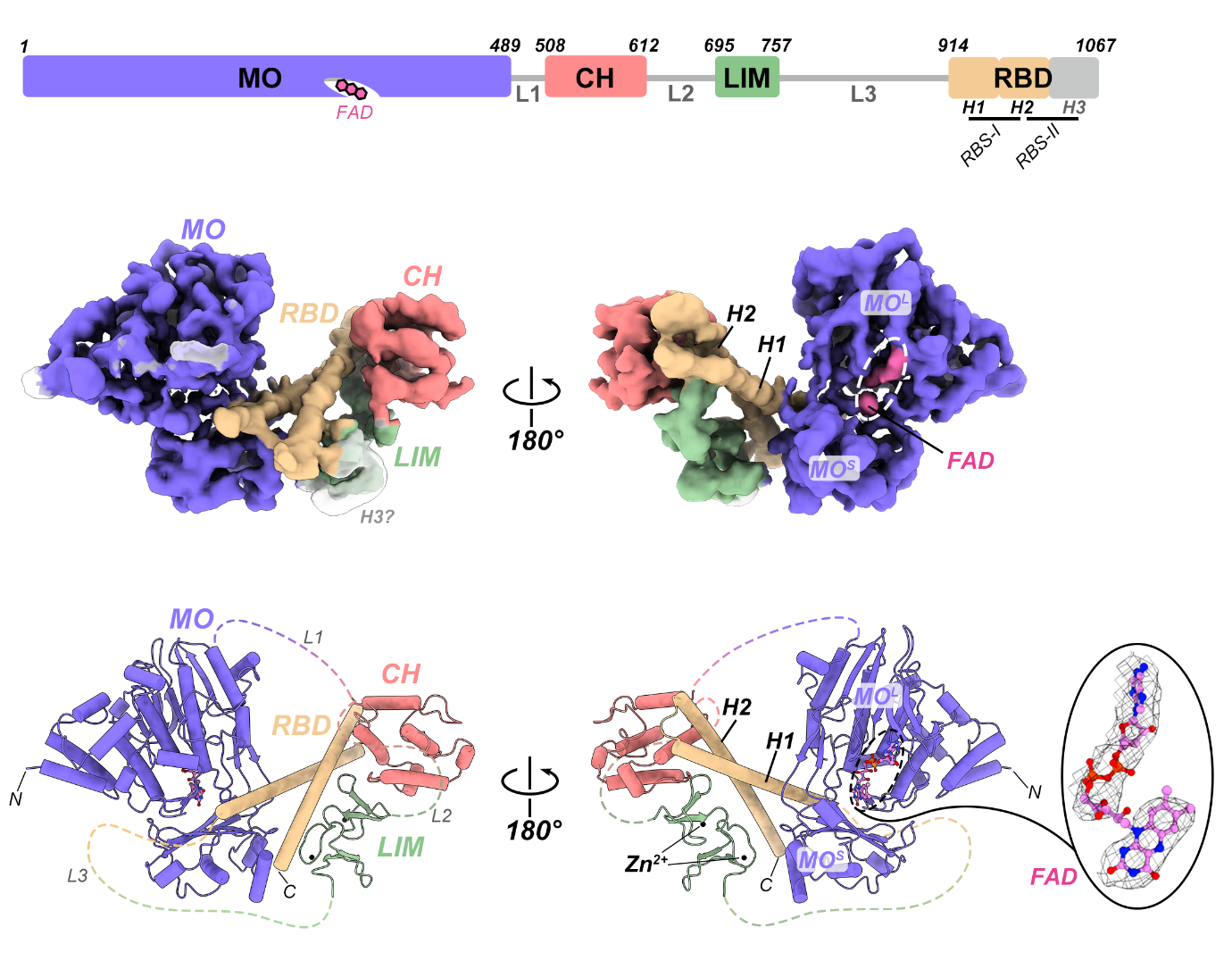
Figure 1. Cryo-EM structure of the full-length MICAL1 protein in its autoinhibited state
The research team found that the N-terminal monooxygenase (MO) domain of MICAL1 and the C-terminal CH-LIM-RBD region assemble together through head-to-tail interaction to build the autoinhibitory conformation. In this autoinhibited structure, the three domains, including CH, LIM, and RBD at the tail region, form a compact ternary structure, modifying the conformation and orientation of the very C-terminal RBD to enable to directly bind to the N-terminal MO domain. This binding of the RBD stabilizes the conformational dynamics of the flexible loops surrounding the active site of the MO domain, which prevents the flipping of the isoalloxazine ring of the FAD cofactor at this active site during catalysis, consequently inhibiting the monooxygenase activity of the MO domain (Fig. 2).
The binding of the CH-LIM-RBD region on MO can also prevent the recognition of the MO domain to its substrate, F-actin. Thus, in the autoinhibited conformation, MICAL1 suppresses its enzymatic activity by blocking not only its catalytic reaction but also the substrate binding through its head-to-tail interaction.
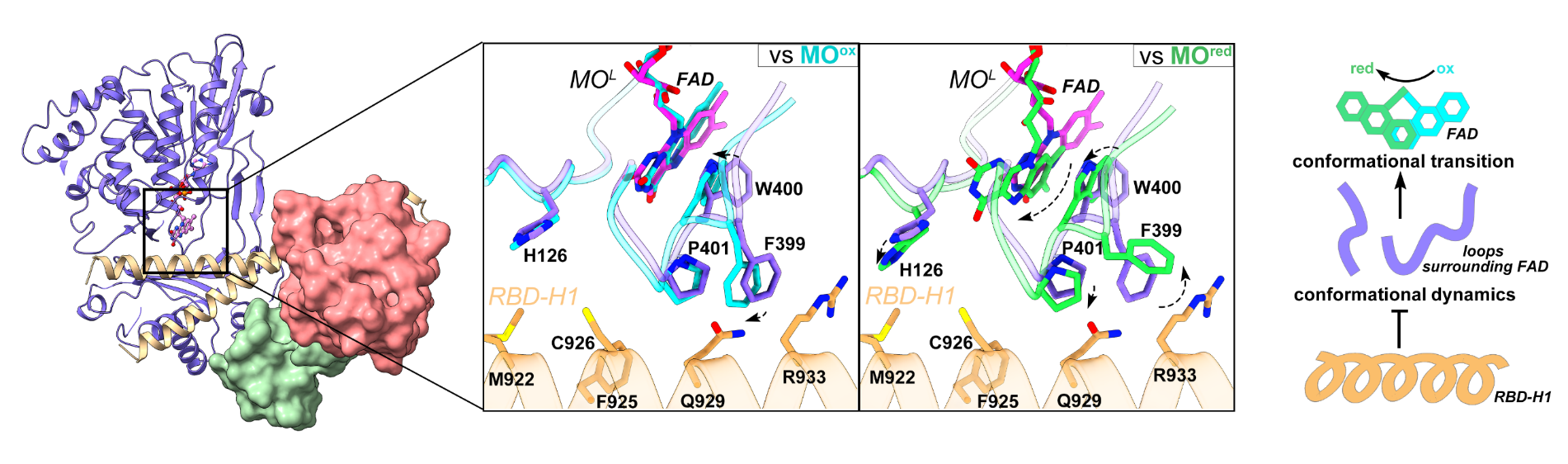
Figure 2. The head-to-tail interaction in MICAL1 facilitates the inhibition of its N-terminal MO domain enzymatic activity
Further analysis revealed how the autoinhibition is relieved by the binding of Rab effectors, which are a classical family of small GTPases expressed universally in cells. The RBD domain at the very C-terminus of MICAL1 owns two Rab-binding sites (RBS-I and RBS-II), which have previously been found to be crucial for recruiting Rab effectors to activate MICAL proteins. Through a series of structural analyses and biochemical validations, the researchers proposed a multi-step autoinhibition relief model (Fig. 3). In this model, the first Rab molecule binds to the RBS-II site on the RBD, stabilizing the helical conformation of its third helix (H3), which consequently disrupts the direct binding between the RBD and CH domains, thereby to expose the RBS-I site on the RBD.
The second Rab molecule binds to the RBD via the RBS-I, further disrupting the interaction between the CH-LIM domains and the RBD and simultaneously changing the orientation of the first helix (H1), which results in the dissociation of the RBD from the MO domain. Finally, the MICAL1 autoinhibition is relieved to open conformation, and the unrestricted MO domain is allowed to enzymatically oxidize and depolymerize the actin cytoskeleton.
To verify whether MICAL family proteins employ a conserved mechanism for regulating activity, the team also investigated another member, MICAL3. With amino acid sequence analysis and mutagenesis study, MICAL3 is confirmed to also form a head-to-tail autoinhibited conformation. Meanwhile, due to the high conversation of the key residues on the RBS-I and RBS-II among MICAL family proteins, the study suggests that MICAL3 likely shares the same Rab-mediated activation mechanism as MICAL1. Therefore, MICAL family proteins generally regulate their oxidase activity through a head-to-tail mediated autoinhibition, which is strictly controlled. This autoinhibition is relieved in multiple steps by the dual binding of Rab effectors (Fig. 3), thereby locally facilitating the cellular dynamics of the actin cytoskeleton and regulating diverse cell activities involved in the cytoskeleton.
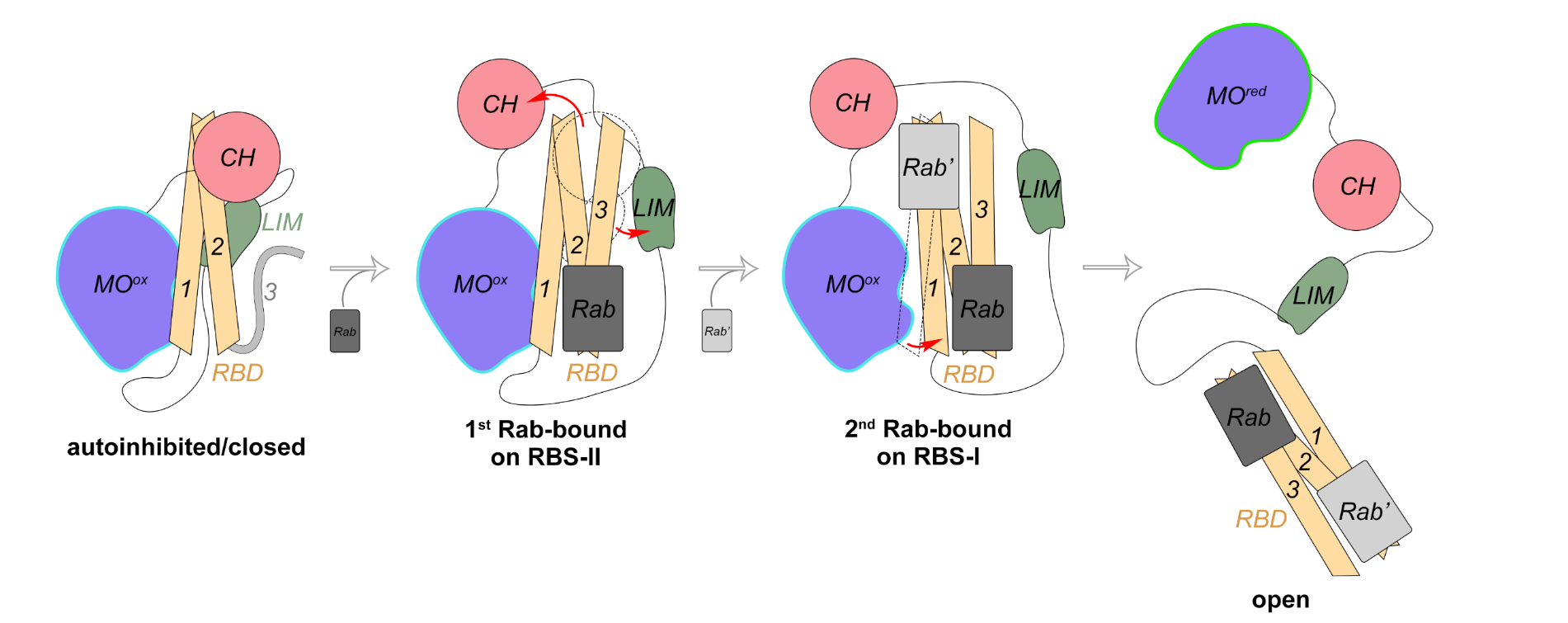
Figure 3. Proposed model of the autoinhibition and relief of MICAL family proteins
Ph.D. student Leishu Lin and master’s student Jiayuan Dong from the School of Life Sciences at SUSTech are the co-first authors of the paper. Associate Professor Zhiyi Wei and Research Associate Professor Fengfeng Niu are the corresponding authors.
This work was supported by the Key-Area Research and Development Program of Guangdong Province, National Natural Science Foundation of China (NSFC), Guangdong Basic and Applied Basic Research Foundation, Shenzhen Science and Technology Program, Shenzhen Key Laboratory of Biomolecular Assembling and Regulation, and the Cryo-EM Center and Core Research Facilities of SUSTech.
Paper link: https://www.nature.com/articles/s41467-024-50940-7
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo ByDepartment of Neuroscience