The CRISPR-Cas system has wide applications in genetic editing, transcriptional, and translational regulation. Based on the CRISPR-Cas system, a variety of CRISPR activation (CRISPRa) transcriptional activation tools have been developed through the fusion of diverse transcriptional activation elements. These systems enable the specific modulation of target gene expression without causing genomic damage or mutations, positioning them as promising tools for gene therapy applications.
Highly efficient CRISPRa transcriptional activation systems often require multiple transcriptional activation elements, rendering them challenging for delivery through the AAV viral system and significantly impeding their utility in gene therapy. In recent years, the functional role of phase separation in transcriptional regulation has garnered increasing attention. Intrinsically disordered regions (IDRs) have been recognized to enhance gene transcription activation in diverse transcriptional regulatory components, thus unveiling a new pathway for crafting a robust CRISPRa transcriptional activation system.
Chair Professor Wei Chen’s lab at the School of Life Sciences, Southern University of Science and Technology, has published a study on the development of an efficient CRISPRa system based on IDR and Modular domain (MD) and elucidated the regulatory mechanisms of transcription through multivalent interactions in the biological process.
Their work, entitled “Specific multivalent molecules boost CRISPR-mediated transcriptional activation”, has been published in Nature Communications.
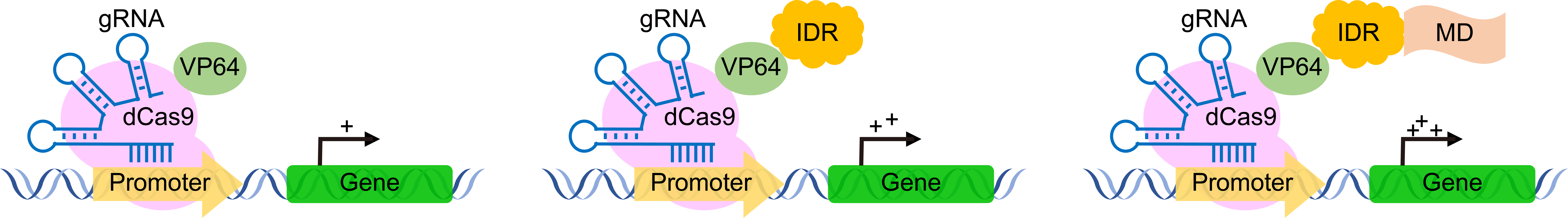
Figure 1. CRISPRa based on dCas9-VP64-IDR/MD
In this study, the researchers fused dCas9-VP64 with various IDRs and assessed the impact of IDRs on the transcriptional activation of the fused CRISPRa protein dCas9-VP64 (Figure 1). They discovered that out of the 12 IDRs tested, seven were capable of enhancing dCas9-VP64 transcriptional activation, with dCas9-VP64-FUS demonstrating robust activation across different target genes. Consequently, they focused their subsequent investigations on this fusion protein and further validated its transcriptional activation specificity.
By studying mutants with defective multivalent interactions in the FUS IDR, the lab found that these interactions are crucial for enhancing transcriptional activation. However, live-cell imaging and fluorescence recovery after photobleaching experiments on effective dCas9-VP64-FUS and non-effective dCas9-VP64-TDP43 revealed that the enhancement of transcriptional activation was not coupled to liquid-liquid phase separation. Further immunoprecipitation experiments unveiled that both dCas9-VP64-FUS and dCas9-VP64-TDP43 could recruit the core protein BRG1 of the BAF chromatin remodeling complex. Yet, only dCas9-VP64-FUS was able to recruit RNA polymerase II and enhance transcription.
MD is another multivalent molecule capable of mediating phase separation, although previous studies did not associate it with transcriptional activation. The researchers found that directly fusing MD with dCas9-VP64 had no significant impact on its transcriptional activation. However, when fused MD with dCas9-VP64-FUS, it substantially enhanced the transcriptional activation capacity of the latter (Figure 1), indicating that the combination of IDR and MD can cooperatively enhance the effect of transcriptional activation. By utilizing dCas9-VP64-IDR/MD activators in transcriptional activation experiments on reporter genes containing varying numbers of repeat target sequences, they further discovered that an excess of cis-trans interactions would repress transcriptional activation and robust transcriptional activation requires optimal cis-trans interactions.
To augment the transcriptional activation effect of the CRISPRa transcriptional activation system on gene transcription further, they targeted the fused CRISPRa transcriptional activation proteins to gene enhancers and promoters simultaneously. They discovered that targeting enhancer-promoter pairs led to a significant enhancement in transcriptional activation efficiency. However, similar outcomes were not observed with dCas9-VP64, implying an essential role for IDR/MD in mediating enhancer-promoter synergy.
Lastly, an artificial enhancer-promoter was constructed and looped through the ABI1-PYL1 chemical induction system. The researchers found that it further enhanced transcriptional activation mediated by the CRISPRa system (Figure 2). Therefore, this study not only developed an efficient gene transcription activation platform but also provided crucial insights into the field of gene transcriptional regulation.
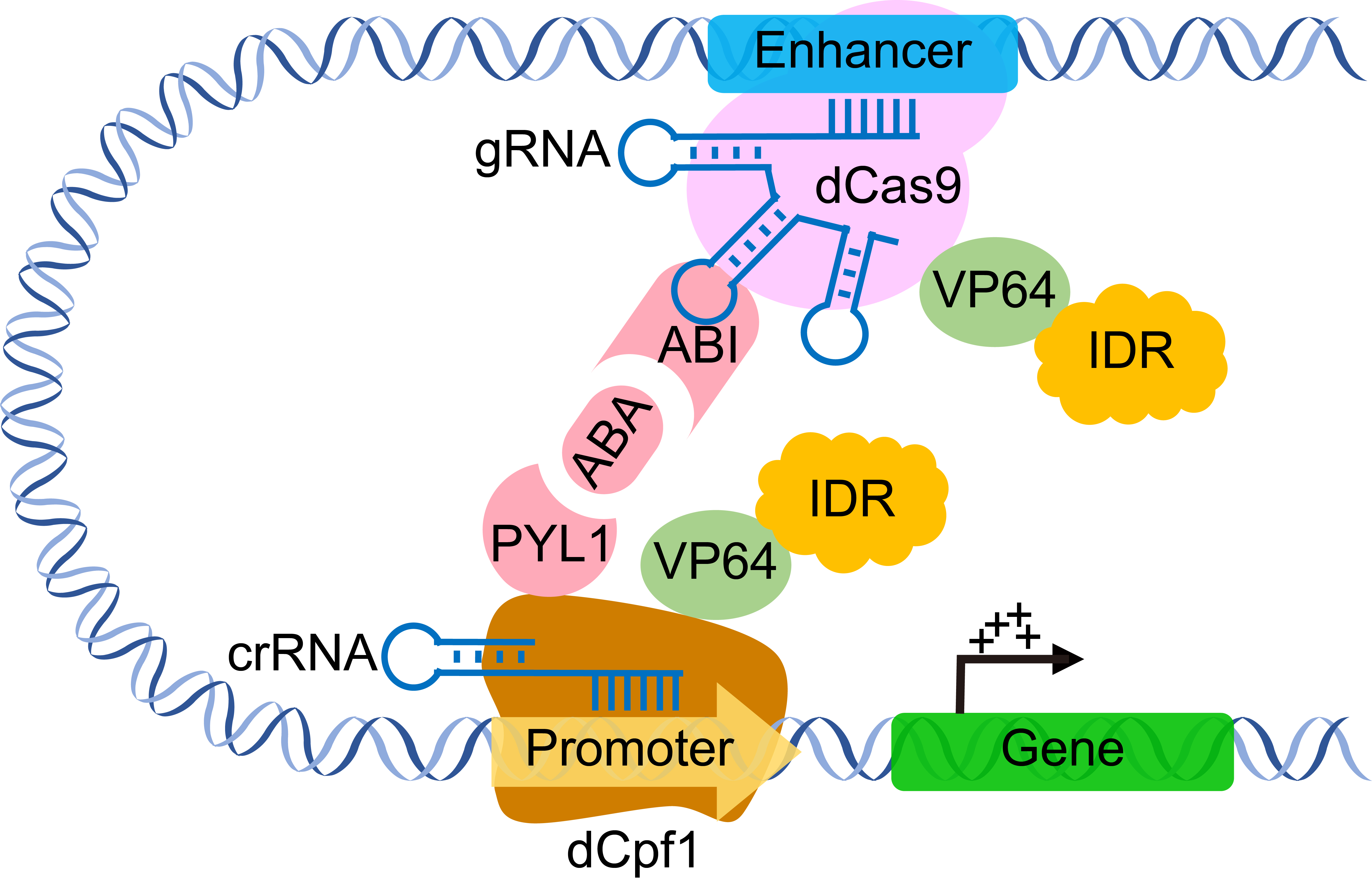
Figure 2. Chromatin interaction enhances CRISPRa-mediated transcriptional activation
Doctoral candidate Rui Chen from the School of Life Sciences at SUSTech is the first author of the paper. Chair Professor Wei Chen and Research Associate Professor Huanhuan Cui are the corresponding authors, and SUSTech is the first affiliated unit. Other contributor to this work included Assistant Professor Meizhen Zheng’s lab, Chair Professor Mingjie Zhang, and Dr. Guanhua Bai, all of SUSTech.
This research was supported by the National Key R&D Program, Shenzhen Science and Technology Innovation Commission, and the Shenzhen Key Laboratory of Gene Regulation and Systems Biology.
Paper link: https://www.nature.com/articles/s41467-024-51694-y
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo ByChen Wei Lab