Tuberculosis (TB) is a global infectious disease primarily caused by Mycobacterium tuberculosis (MTB). In recent years, over 10 million new TB cases have emerged annually, leading to over 1.6 million deaths, making MTB the second deadliest infectious pathogen globally, surpassed only by SARS-CoV-2 and ahead of HIV. The emergence and increasing prevalence of drug-resistant TB have exacerbated the global TB epidemic.
There are two primary mechanisms of MTB drug resistance: mutations in drug target-related genes rendering drugs ineffective, and the activation and overexpression of MTB drug efflux pumps, which decrease the effective drug concentration within bacteria. Efflux Pump A (EfpA) is the first drug efflux pump discovered in MTB and is essential for MTB survival. In various clinical drug-resistant strains, overexpression of EfpA increases MTB’s tolerance to drugs like isoniazid, rifampicin, and amikacin by tens to hundreds of fold. Recently, novel anti-MTB inhibitors, the BRD-8000 series and BRD-9327, have been shown to specifically target EfpA. However, our understanding of EfpA’s function and inhibition mechanisms within MTB remains limited.
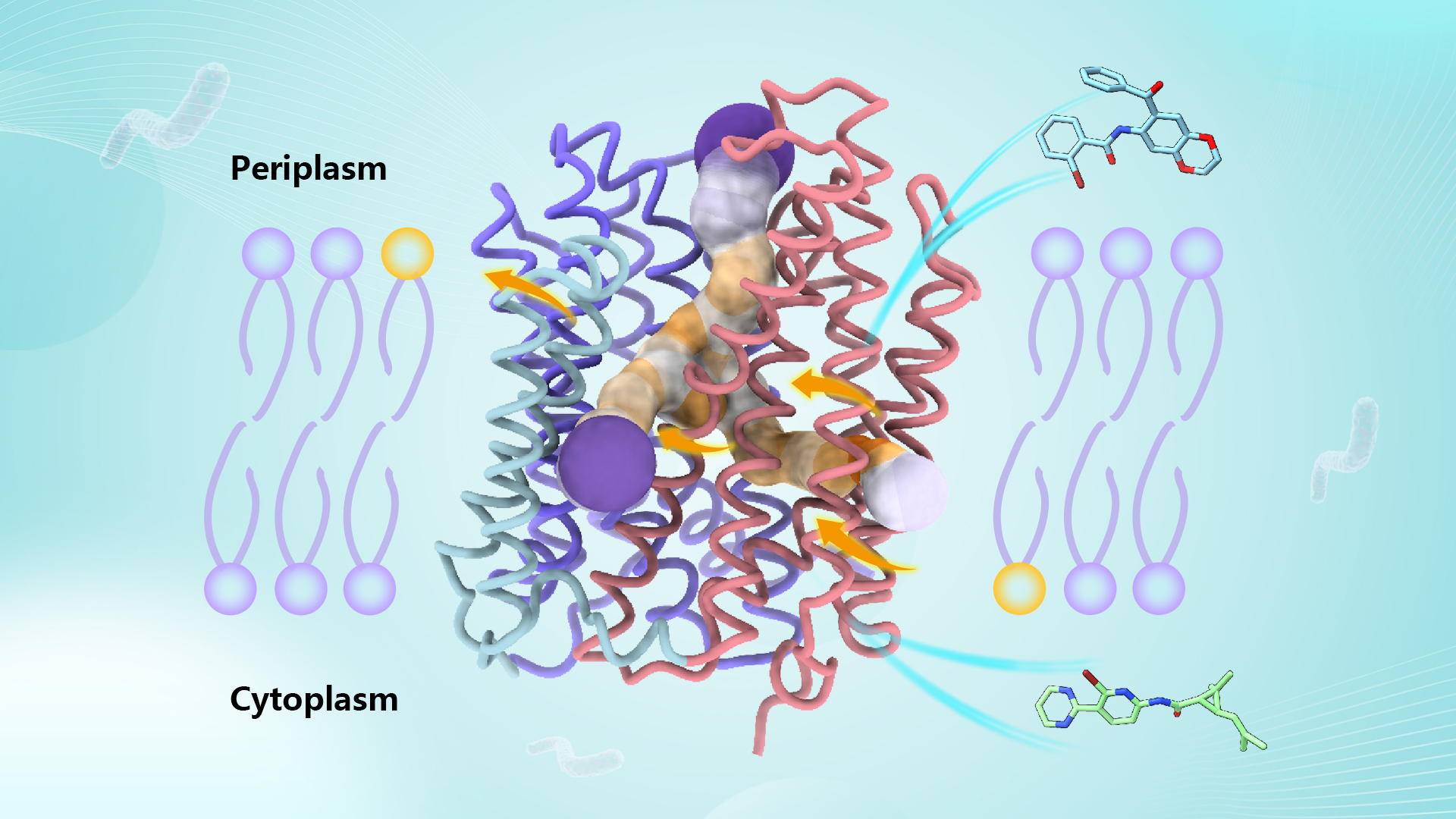
A research team led by Chair Professor Maofu Liao from the School of Life Sciences at the Southern University of Science and Technology (SUSTech) has published a study that suggests that EfpA may function as a lipid transporter, facilitating lipid flip-flop between phospholipid bilayers within MTB cells. It also reveals the binding sites and inhibition mechanisms of novel inhibitors targeting EfpA.
Their work, entitled “Structures of the Mycobacterium tuberculosis efflux pump EfpA reveal the mechanisms of transport and inhibition”, was published in Nature Communications on September 4, 2024.
The research team utilized cryo-electron microscopy to resolve high-resolution structures of EfpA in an outward-open conformation bound with endogenous lipids or inhibitors. EfpA belongs to the Major Facilitator Superfamily (MFS) of transporters, but unlike classical MFS transporters with 12 transmembrane helices, EfpA features six transmembrane helices in each of its N-terminal domain (NTD) and C-terminal domain (CTD), along with a hinge domain (HD) with two additional transmembrane helices connecting NTD and CTD.
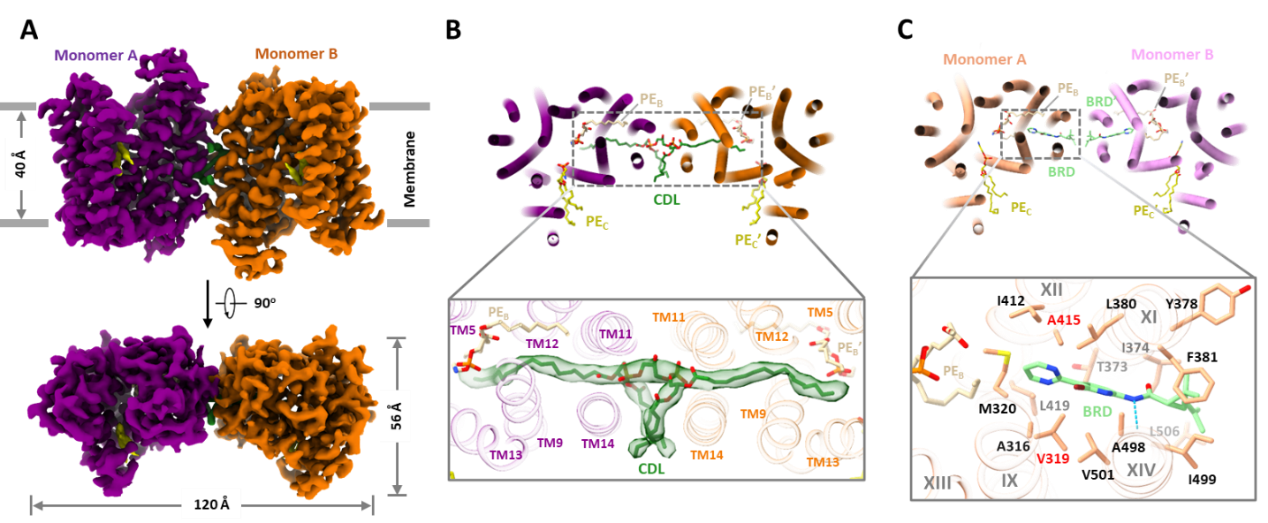
Figure 1. Structure of Mycobacterium tuberculosis EfpA, lipid channel, and inhibitor binding sites
Intriguingly, EfpA harbors a “staircase”-like channel extending from the inner leaflet to the outer leaflet of the plasma membrane phospholipid bilayer, where three lipid molecules bind sequentially in a head-to-head, tail-to-tail manner (positions A, B, and C). Molecular dynamics simulations indicate that these lipid molecules stably bind within this channel. In the central region of the channel, the hydrophilic heads of two lipid molecules, along with surrounding helical structures, form an outward-open negatively charged pocket whose size and surface charge match the hydrophilic substrates EtBr and isoniazid effluxed by EfpA.
In the EfpA-BRD-8000.3 complex structure, BRD-8000.3 replaces the lipid molecule at position A, occupying the inner leaflet lipid binding site within EfpA’s lipid channel. This is consistent with previously reported mutation sites in several BRD-8000.3-tolerant mutants. In contrast, BRD-9327 tolerance mutations localize around lipid binding site B. AutoDock molecular docking analysis shows that BRD-9327 can bind to lipid binding site B, displacing the lipid there. The distinct binding sites of BRD-9327 and BRD-8000.3 in EfpA explain their synergistic inhibitory effect when combined.
Small molecules occupying lipid binding sites in EfpA, thereby inhibiting MTB, indicate a crucial link between these lipids and EfpA’s intrinsic function in MTB. To further explore EfpA’s function, the researchers employed the gene structure-function prediction tools COFACTOR and Dali to search databases, both consistently identifying lysophospholipid transporter MFSD2A in the MFS superfamily as the best match. Despite low sequence homology (11%), EfpA and MFSD2A exhibit structural similarities. Furthermore, lipid binding sites B and C also correspond. These findings suggest that EfpA likely functions as a lipid transporter, similar to MFSD2A.
In addition, the researchers compared the cryo-EM structure of EfpA in an outward-facing open conformation with the inward-facing open conformation predicted by AlphaFold, revealing conformational changes and alterations in the lipid channel between the two states. Ultimately, they proposed a “staircase-flip” model for EfpA’s lipid transport mechanism, as well as the mechanism of action for small molecule inhibitors.
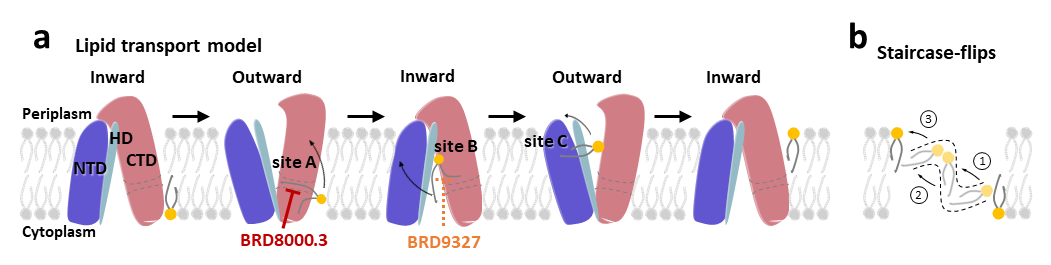
Figure 2. The lipid transport model and small molecule inhibition mechanism of EfpA
In summary, this study has elucidated the structure of the multidrug efflux pump EfpA in Mycobacterium tuberculosis, suggesting that EfpA may function as a lipid transporter in MTB. It has identified the binding sites of novel MTB inhibitors within EfpA. These advancements provide a structural basis for the research and optimization of novel anti-tuberculosis
Dr. Shuhui Wang, formerly a postdoctoral fellow at Harvard University and now at Yale University, and Chair Professor Maofu Liao have shed light on the lipid transport model and small molecule inhibition mechanism of the Mycobacterium tuberculosis efflux pump EfpA.
Dr. Shuhui Wang is the first and co-corresponding author of the paper, with Chair Professor Maofu Liao serving as a co-corresponding author. This work was supported by the National Natural Science Foundation of China (NSFC).
Paper link: https://www.nature.com/articles/s41467-024-51948-9
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByYingying XIA
Photo BySchool of Life Sciences