The growth in data storage and computing has long driven the microelectronics and integrated circuit industry. In the era of big data, the massive increase in data generation challenges current storage systems.
DNA, with its remarkable storage density—capable of holding hundreds of exabytes per gram and remaining stable for thousands of years—offers a solution that surpasses traditional media. Leading organizations like Microsoft and Western Digital have formed the DNA Data Storage Alliance to propel this innovative technology forward. However, conventional DNA sequencing methods suffer from slow data retrieval, often requiring days to access stored information, which significantly limits their potential use in real-time applications.
Nanopore sequencing, in contrast, emerges as a promising alternative, capable of analyzing DNA sequences in real-time by detecting changes in electrical current as individual molecules pass through a nanopore.
This approach enables fast sequence readout, ideal for high-speed data storage. However, nanopore technology faces a high insertion-deletion error rate, a key challenge for DNA data storage. Improving encoding and decoding algorithms to address these errors is essential for more reliable DNA-based storage.

Associate Professor Yi Li’s research group from the School of Microelectronics at the Southern University of Science and Technology (SUSTech) has made notable advancements in the encoding-decoding field of DNA storage. Their encoding and decoding method successfully achieves rapid readout of DNA storage data under high insertion and deletion errors.
Their work, titled “Composite Hedges Nanopores Codec System for Rapid and Portable DNA Data Readout with High INDEL-Correction”, has been published in the multidisciplinary journal Nature Communications. It was also selected as a featured article in the Editors’ Highlights section of the journal.
Based on the above-mentioned contexts, Yi Li’s group introduced a codec system tailored for nanopore sequencing, named the Composite Hedges Nanopores (CHN) codec. This system significantly enhances error correction capabilities, enabling effective data recovery in high-error environments.
This approach facilitates the application of nanopore sequencing for data storage and paves the way for portable, efficient data readout even in extreme environments. As a result, the CHN codec represents a promising advancement in information storage and data security for future applications.
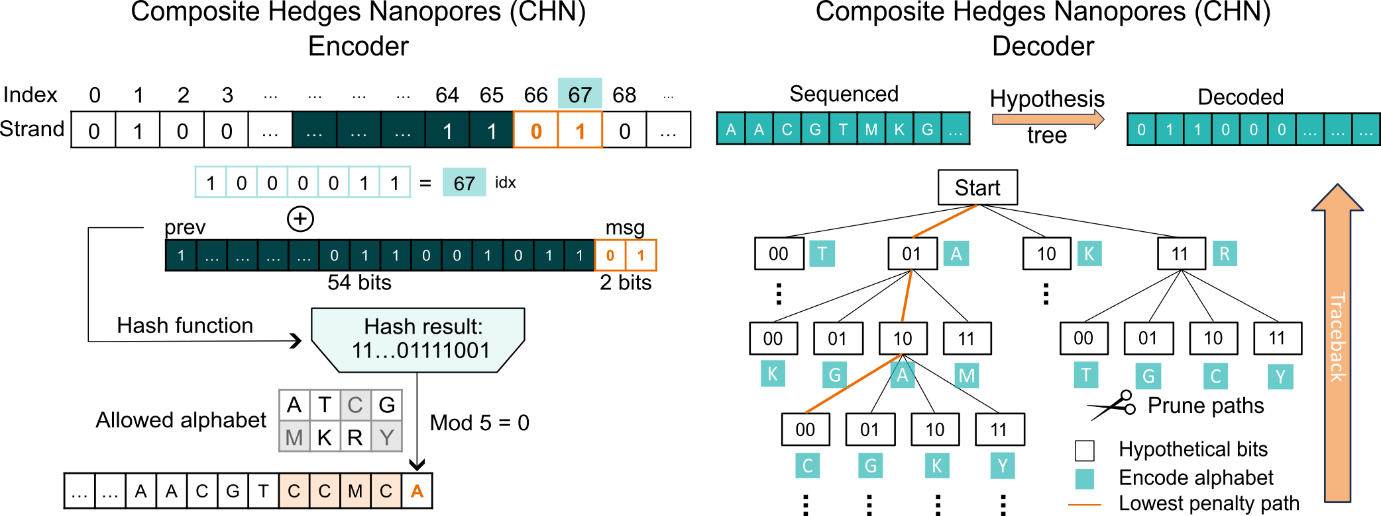
Figure 1. CHN codec architecture for high insertion/deletion error correction
The workflow of the CHN encoding scheme and its performance within the nanopore sequencing system are shown in the system’s complete workflow (Figure 1). By employing methods such as degenerate bases, anchor sequence embedding, and constraint filtering, this encoding process increases the tolerance of the DNA storage system to indel errors, effectively reducing the risk of data loss. Furthermore, comparisons with other encoding strategies highlight CHN’s significant advantages in error tolerance and data recovery rates, demonstrating its robustness in high-error conditions.
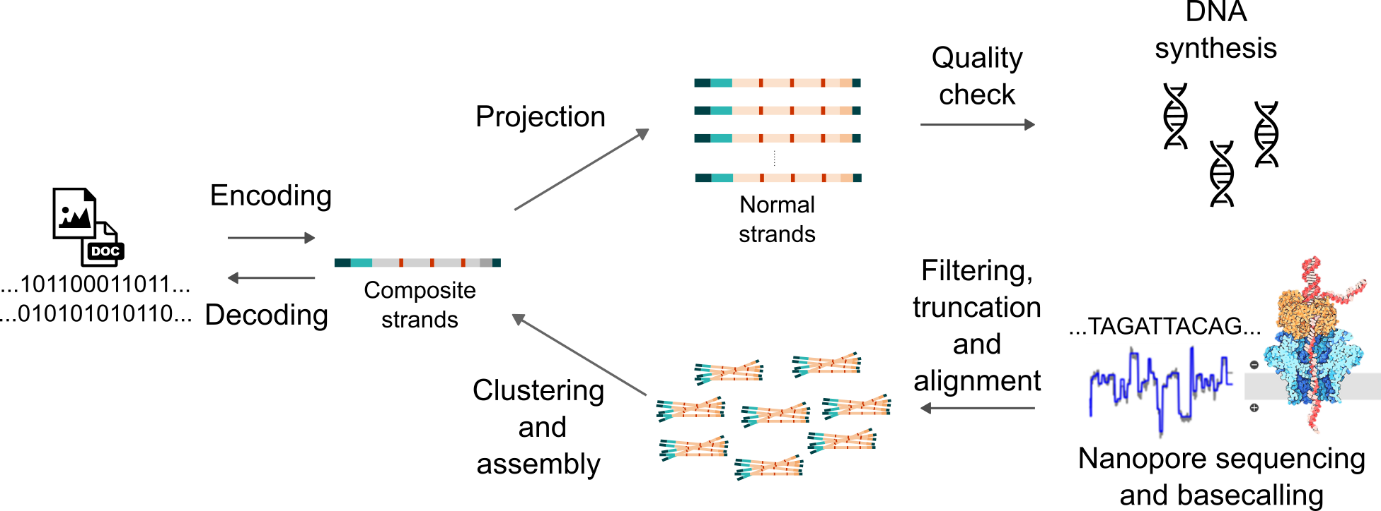
Figure 2. Simulation analysis of DNA data recovery rates under various error rates
Binary data recovery rates under varying indel and substitution error rates show that the CHN encoding system maintains a 100% data recovery rate, even at indel rates as high as 16% (Figure 2).
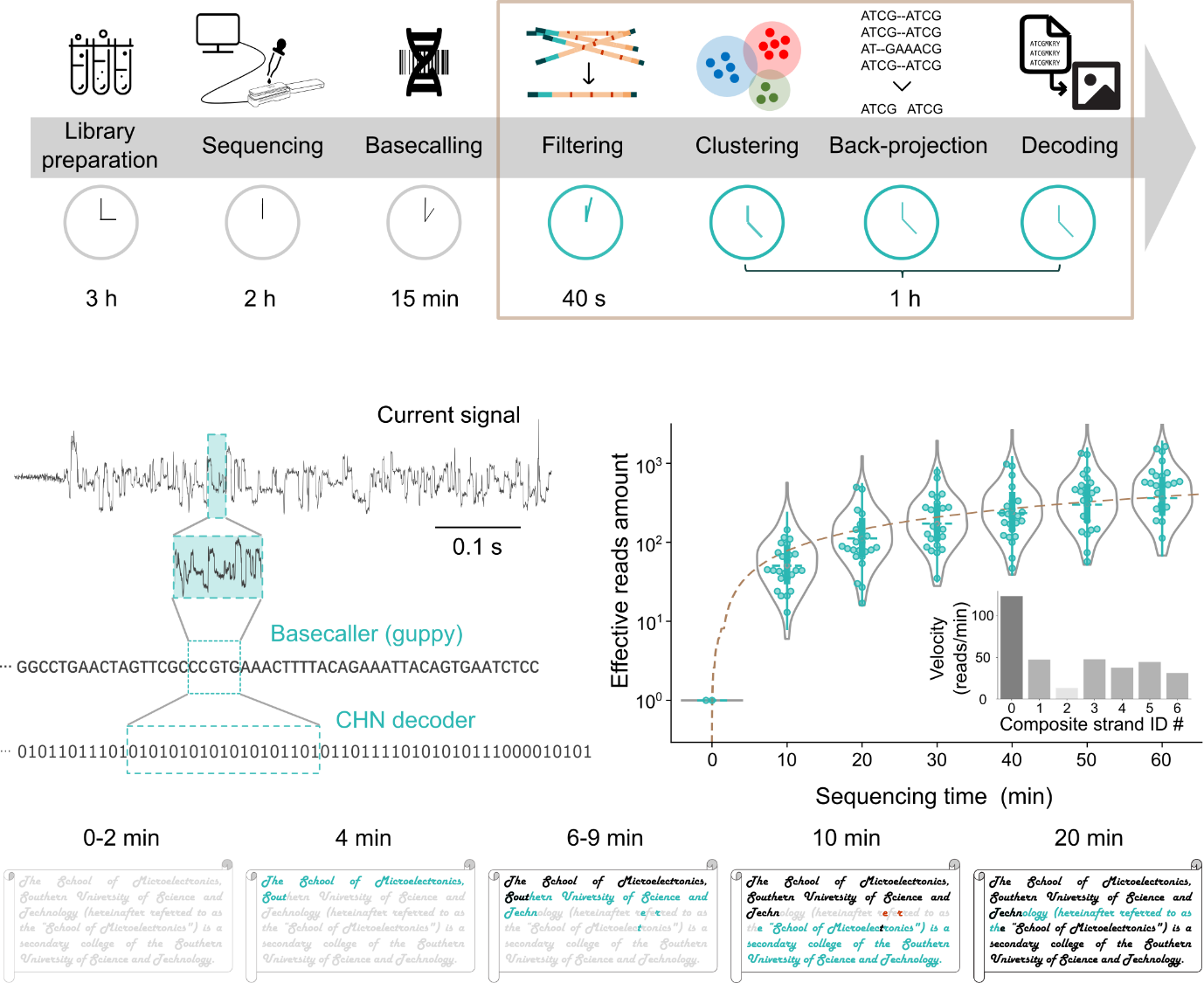
Figure 3. Text data retrieval based on CHN encoding
The results of an in vitro experiment for text data recovery based on the CHN encoding scheme show that a text file encoded into seven composite strands was completely recovered within 20 minutes. This substantial reduction in recovery time advances the practical implementation of DNA data storage technology. Additionally, the experiment verifies the efficiency of CHN encoding through the effective distribution of nanopore reads (Figure 3).

Figure 4. CHN-based image file recovery and performance analysis
In another in vitro test, an image file was successfully recovered within 120 minutes, demonstrating CHN’s strong recovery capabilities for larger data files. Although the recovery time for image files is slightly longer than for text files, the CHN system still exhibits excellent data integrity even at lower coverage, opening new pathways for large-scale data storage in DNA (Figure 4).
Xuyang Zhao and Junyao Li, both master’s graduates and current Ph.D. candidates in the School of Microelectronics at SUSTech, are the first co-authors of the paper. Associate Professor Yi Li and Professor Qing Pan from Zhejiang University of Technology are the co-corresponding authors. Other contributors to this work from SUSTech include Professor Jixian Zhai, Research Associate Professors Yanping Long and Ronghui Liu, master’s student Mr. Qingyuan Fan, and visiting master’s graduate Jing Dai.
Paper link: https://doi.org/10.1038/s41467-024-53455-3
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo BySchool of Microelectronics