Plant senescence is a highly orchestrated biological process that involves systematic gene regulation, as well as the nutrient transport and allocation across multiple cells, tissues, and organs. This process is vital for plant growth, reproduction, and adaptation to dynamic environments. During senescence, plants break down organelles and macromolecules such as chloroplasts and proteins, translocating essential nutrients like carbon and nitrogen from source organs (leaves) to sink organs (roots, flowers, fruits, and seeds) to sustain growth and reproduction. The onset and progression of senescence are regulated by diverse internal and external factors, including developmental stages, hormones, and environmental stresses.
The timely initiation of senescence is evolutionally linked to adaptation to complex environments, while appropriately delaying senescence can significantly enhance crop yield and quality. Although numerous senescence-associated genes (SAGs) have been studied across species, fundamental questions remain unsolved: How do plants integrate developmental stage and environmental signals to trigger senescence? How do plants coordinate regulatory programs of senescence and nutrient allocation across organs and cell types? Addressing these questions by elucidating key regulatory networks and identifying critical SAGs will enable precise manipulation of plant growth and senescence processes. This, in turn, could promote crop productivity and quality.

The research team led by Chair Professor Hongwei Guo from the Department of Biology at the Southern University of Science and Technology (SUSTech), in collaboration with Wuhan BGI-Research Institute, has made a significant advancement in understanding the cellular and molecular mechanisms of leaf senescence. They constructed a comprehensive single-nucleus transcriptome atlas spanning the entire life cycle of the model organism Arabidopsis thaliana. By formulating the Senescence-Associated-Gene index (SAG-index) and Youth-Associated-Gene index (YAG-index), they innovatively elucidated the leaf senescence process at single-cell resolution, and described the scenario of dynamic carbon and nitrogen allocation from source to sink.
Their groundbreaking study, entitled “An Arabidopsis single-nucleus atlas decodes leaf senescence and nutrient allocation”, has been published in Cell on April 11, 2025.
The research team employed single-nucleus transcriptome sequencing (snRNA-seq) technology to analyze 20 major tissues spanning the entire life cycle of Arabidopsis, constructing a high-resolution atlas encompassing over one million nuclei. By integrating published genetic marker information, they systematically annotated the 539 cell clusters identified across individual tissues and multiple developmental stages, resulting in 38 distinct cell types. Based on this atlas, they elucidated developmental trajectories of critical cell types through cross-stage cell comparisons and hub genes’ expression dynamics. They also constructed the cross-organ conserved and organ-specific transcription factor regulatory networks. This high-quality single-nucleus transcriptome atlas provides a foundational reference and invaluable resource for future studies on plants.
Focusing on the leaves across six consecutive developmental stages, the researchers discovered that the three major cell types in leaves—epidermal cells (EPI), mesophyll cells (MES), and vascular cells (VAS)—exhibit systemic coordination yet intercellular heterogeneity during senescence. Leveraging these insights, two indexes—the SAG-index and the YAG-index—were formulated to estimate cellular senescence state at the single-cell level. SAG-index and YAG-index were calculated based on the expression profiles of SAGs highly expressed at late-senescence stages and YAGs predominantly expressed at early developmental stages, respectively.
With these two indexes, they successfully quantified the senescence states at single-cell resolution and delineated the distinct stages of senescence onset and progression. Through analyzing published bulked RNA-seq data, these indexes were able to track multi-organ development, cellular differentiation, and stress responses in both Arabidopsis and other plant species. These breakthroughs elevated plant senescence research to a single-cell level and laid a solid foundation for multidimensional molecular dissection.
Through systematic co-expression network analysis, the team resolved the senescence regulatory networks and identified multiple senescence-associated modules with numerous novel SAGs. These newly discovered SAGs exhibited significant co-expression patterns with functionally characterized SAGs, suggesting their potential to synergistically regulate leaf senescence through intricate molecular regulations. Through phenotypic estimation of mutants, chlorophyll quantification, and marker gene expression profiling, the regulatory networks were systematically validated. This senescence-associated regulatory network will provide important genetic resources and a roadmap for senescence-related study.
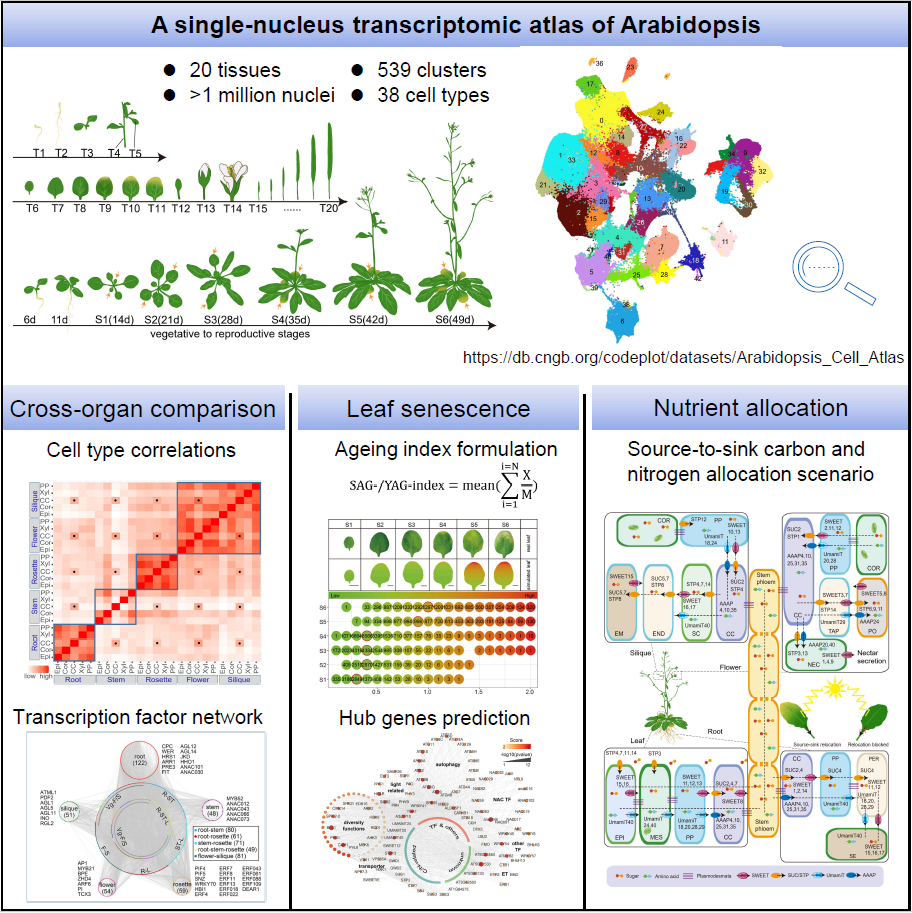
Figure 1. A single-nucleus Arabidopsis atlas decodes leaf senescence and nutrient allocation
Nutrient transport and redistribution during leaf senescence are critical for plant growth and reproduction. Based on the Arabidopsis single-nucleus atlas, the researchers delineated temporo-spatial dynamics by analyzing expression patterns of carbon and nitrogen transporter genes across organs and cell types, with genetic and molecular validation of hub transporters. They found multiple cell-type-specifically expressed transporter genes in both leaves and sink organs. For example, SWEET and SUC family genes were highly expressed in phloem parenchyma (PP) and companion cells (CC), driving sugar export to flowers and siliques. STP transporters in non-CC cells mediate sugar retrieval, while UmamiT genes in senescent PP cells enable amino acid efflux. AAAP transporters in CC cells facilitate apoplastic amino acid uptake. This systematic decoding of cell-type nutrient roadmap uncovers the sophisticated mechanisms underlying carbon and nitrogen reallocation during leaf senescence, providing valuable insights for the understanding of nutrient management.
This study constructed a comprehensive single-nucleus atlas spanning the entire life cycle of Arabidopsis and innovatively promoted senescence-associated research to single-cell resolution. The breakthroughs in this research provide a valuable transcriptomic resource, a multiscale theoretical framework for crop trait engineering, and a new paradigm for plant sciences with single-cell sequencing.
Senior Engineer Yichuan Wang and Research Assistant Professor Wei Yan from Guo’s lab at SUSTech, together with Drs. Xing Guo, Caiyao Zhao, and Cong Tan from Wuhan BGI-Research, are the co-first authors of the paper. Chair Professor Hongwei Guo, along with Xun Xu and Huan Liu from BGI-Research, are the co-corresponding authors. Other contributors to this work include Dan Zhang, Yang Peng, Yuping Qiu, Tao Peng, and Yi Zhang from Guo’s lab.
Paper link: https://doi.org/10.1016/j.cell.2025.03.024
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yilin ZHOU
Photo By