In the long-term evolutionary arms race between bacteria and phages, bacteria have evolved a wide array of anti-phage immune systems to defend against phage invasion. These defense mechanisms are highly diverse, ranging from well-known systems like CRISPR-Cas to lesser-known strategies that are only beginning to be uncovered. Understanding these mechanisms not only sheds light on microbial evolution but also provides valuable insights for biotechnological innovation.

A research team led by Associate Professor Ning Jia from the Department of Biochemistry at the Southern University of Science and Technology (SUSTech) has identified a novel anti-phage defense strategy mediated by the bacterial reverse transcriptase DRT9 and a non-coding RNA. Bacteria produce abundant poly-A-rich single-stranded cDNAs, spanning thousands of nucleotides for anti-phage defense. This discovery expands our understanding of both bacterial anti-phage immunity and reverse transcriptase biological functions, and provides a basis for developing DRT9-based biotechnological tools.
Their paper, titled “Bacterial reverse transcriptase synthesizes long poly-A-rich cDNA for anti-phage defense”, has been published in Science.
To explore the functional mechanisms of the DRT9 immune system, the researchers first expressed the DRT9 system, comprising the reverse transcriptase DRT9 and a non-coding RNA (ncRNA), in E. coli. They demonstrated its ability to resist diverse phages via an abortive infection (Abi) strategy (Figure 1A and 1B). Cryo-EM analysis revealed that DRT9 forms a double-layered hexamer bound to a structured ncRNA with a unique “tree-like” conformation (Figure 1C). Mutational studies further confirmed that the oligomeric state and structural features (rather than sequence) of the ncRNA are critical for its anti-phage function.
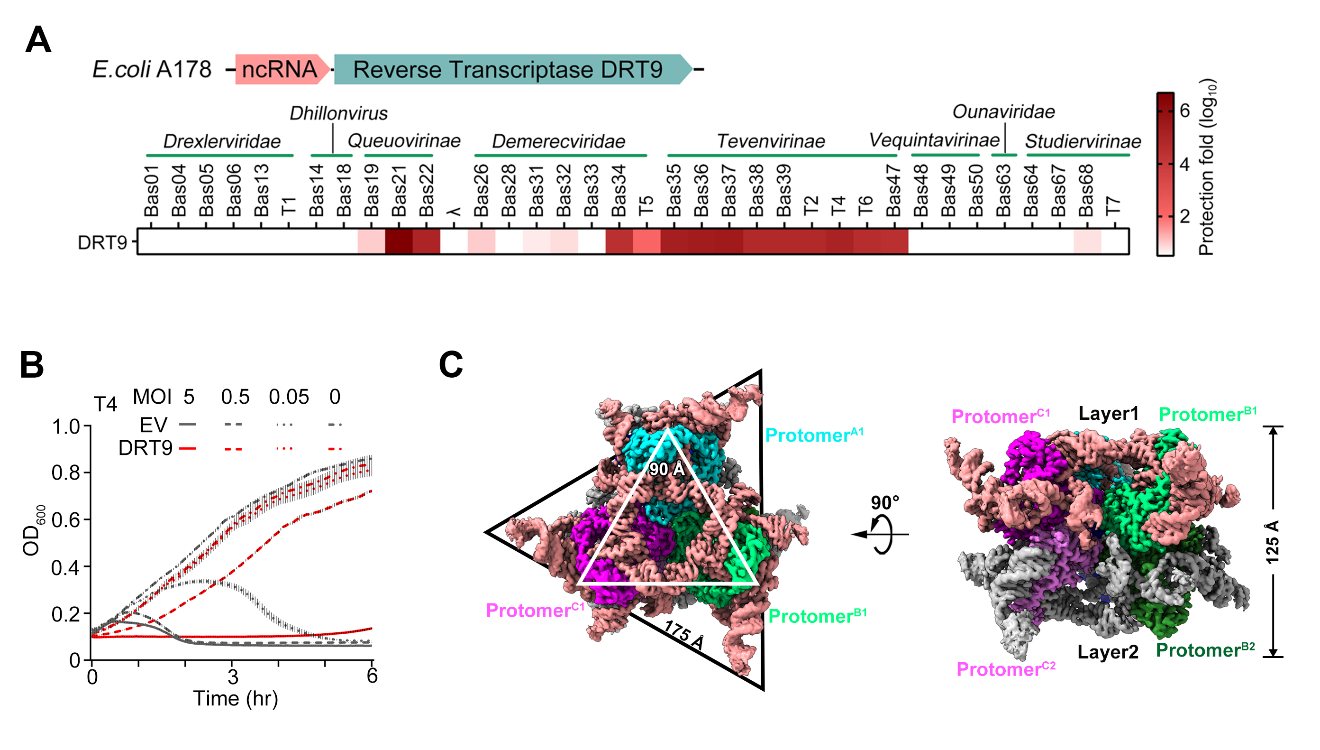
Figure 1. DRT9 assembles into a hexameric complex to resist phage infection
In the presence of high dNTP/Mg²⁺, DRT9 can synthesize single-stranded cDNA longer than 1000 nucleotides without the need for additional primers. Structural insights identified that nucleotide U124 in the ncRNA plays an essential role in cDNA synthesis, while a high concentration of dATP alone is sufficient to trigger cDNA synthesis. Illumina next-generation sequencing (NGS), PacBio single-molecule sequencing in real-time (SMRT) sequencing, together with UPLC-MS and Southern blot analysis, confirmed that the cDNA was composed of poly-A exceeding 1000 nucleotides in length. (Figure 2A-C).
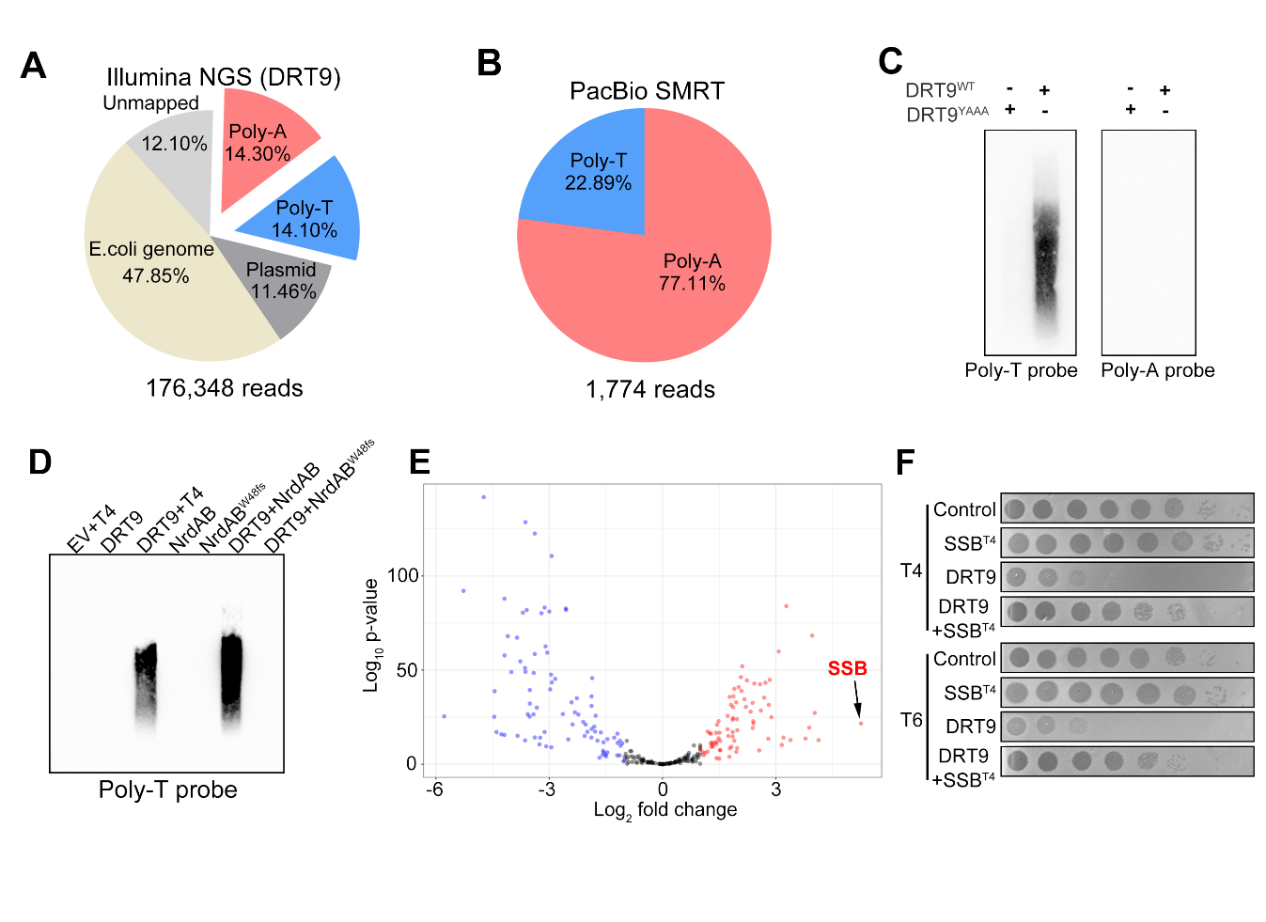
Figure 2. Phage NrdAB activates DRT9 to synthesize long poly-A-rich single-stranded cDNA
To explore how the DRT9-ncRNA complex senses phage invasion, the team isolated phage mutants that escaped DRT9-mediated defense. They identified mutations in nrdA/nrdB (encoding ribonucleotide reductase subunits) that allowed phages to escape DRT9 system. Further experiments showed that phage T4 infection would elevate intracellular dNTP levels by NrdAB, which can activate the DRT9 system. NrdAB mutants failed to increase intracellular dNTP levels, enabling phage escape (Figure 2D). These results indicate that DRT9 is activated by sensing the increased dATP concentration caused by phage infection.
Investigating how the DRT9 system disrupts phage propagation, the researchers performed transcriptomics analysis of cells expressing and not expressing DRT9 during T4 phage infection. This analysis revealed a substantial increase in the expression of genes involved in T4 phage DNA replication, particularly those encoding single-stranded DNA binding (SSB) proteins, during DRT9 activation (Figure 2E). Phage SSB is essential for T4 phage DNA replication; overexpression of phage T4 SSB in E. coli reduced the anti-phage activity of DRT9 and rescued T4 phage replication (Figure 2F). This suggests that the poly-A-rich cDNA synthesized by DRT9 might bind to and sequester phage SSB, thereby inhibiting T4 phage propagation.
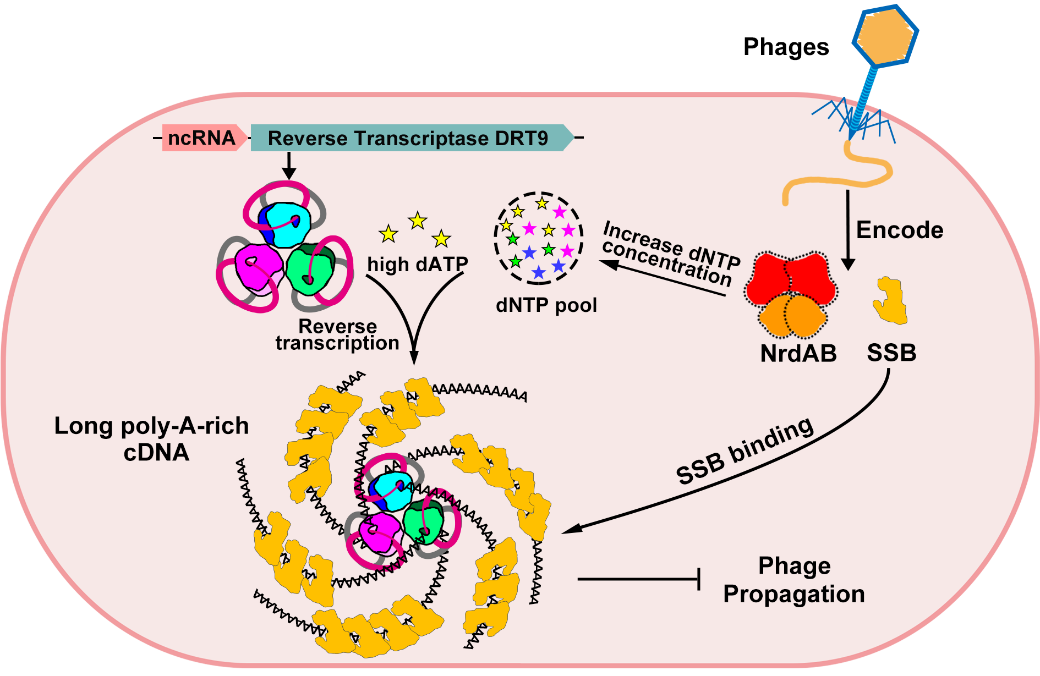
Figure 3. Mechanistic model of DRT9-mediated anti-phage immunity
This study identifies a novel anti-phage immune strategy mediated by bacterial reverse transcriptase DRT9 and non-coding RNA. Upon phage infection, the phage-encoded ribonucleotide reductase NrdAB complex elevates intracellular dNTP levels, preparing for the synthesis of the phage genome. The DRT9 immune system in bacteria exploits this increase in dNTP concentration by sensing the elevated dATP levels, which activate DRT9 to synthesize long, poly-A-rich single-stranded cDNA. This cDNA lacks a secondary structure, making it an ideal substrate for SSB protein binding. The binding of the cDNA to SSB sequesters this key protein involved in phage replication, thereby inhibiting phage replication and protecting the bacteria (Figure 3). This finding enhances our understanding of bacterial immunity, links prokaryotic reverse transcriptases with eukaryotic telomerases, and provides a molecular foundation for the development of DRT-based biotechnological tools.
Ph.D. candidate Xin-Yi Song, along with postdoctorals Yushan Xia and Jun-Tao Zhang from SUSTech, are the co-first authors of this paper. Associate Professor Ning Jia is the corresponding author.
Paper link: http://doi.org/10.1126/science.ads4639
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yuwen ZENG
Photo ByDepartment of Biochemistry