Macroautophagy, commonly referred to as autophagy, is a highly conserved and essential degradation mechanism in eukaryotic cells. When cells are exposed to stress, they initiate autophagy to provide essential materials and energy for survival by non-selectively degrading parts of intracellular components. Simultaneously, autophagy serves to degrade damaged organelles and misfolded proteins, playing a crucial role in cellular material recycling. The dysfunction of autophagy in the nervous system is closely associated with the onset and progression of neurodegenerative diseases.
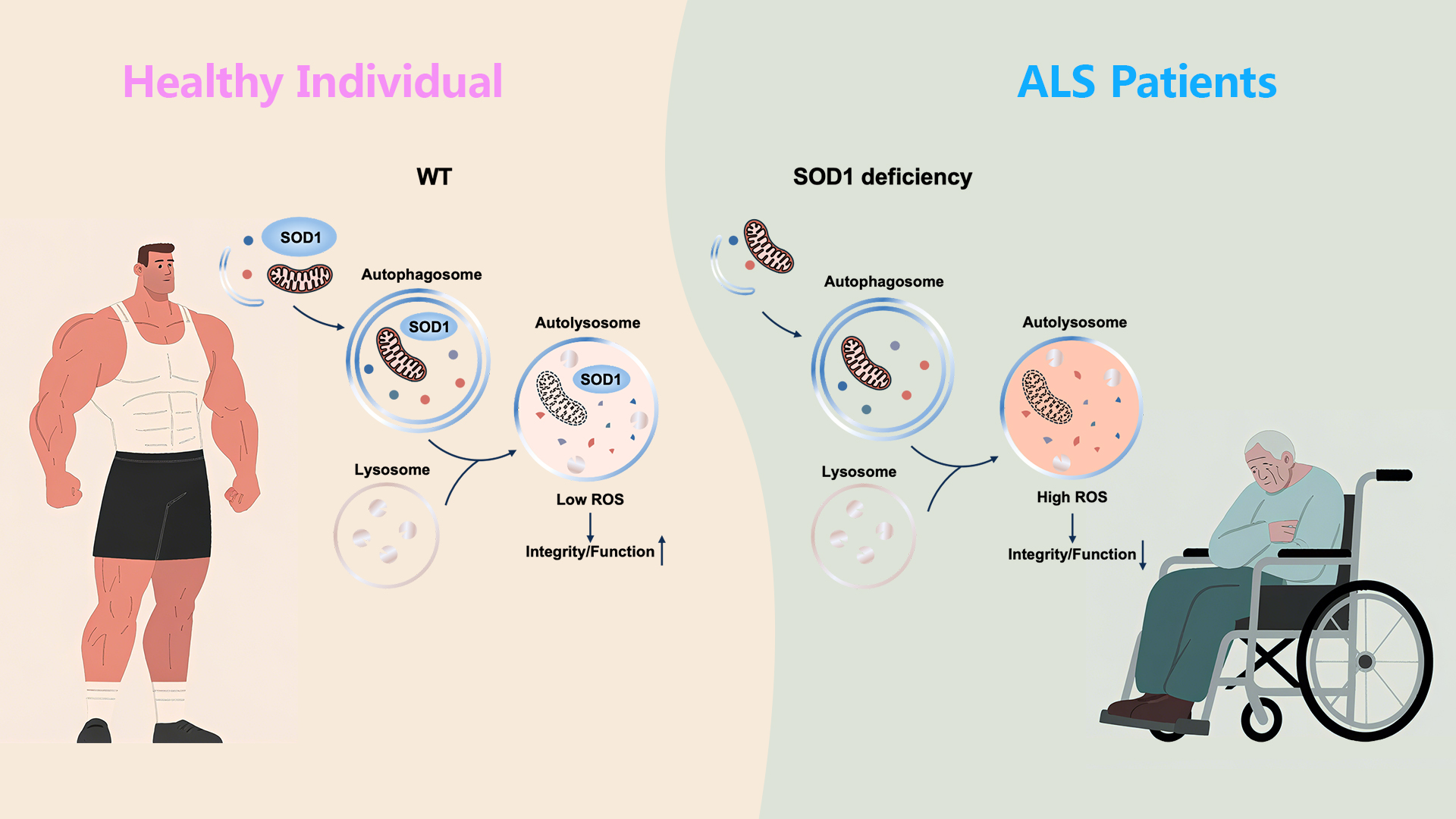
Superoxide dismutase 1 (SOD1) is a critical intracellular antioxidant enzyme. Multiple mutations of SOD1 are involved in the pathogenic cause of amyotrophic lateral sclerosis (ALS). Previous studies report that mutant SOD1 forms protein aggregates and induces cytotoxicity, although the relationship between aggregates and pathogenicity remains debated. These mutations reduce antioxidant activity and disrupt cellular oxidative balance, potentially leading to impaired neural function. Therefore, the pathogenic mechanisms underlying SOD1 mutation-induced ALS remain unclear.
A research team led by Associate Professor Yan Zhao from the School of Life Sciences, in collaboration with Assistant Professor Ruilin Tian from the School of Medicine at the Southern University of Science and Technology (SUSTech), has revealed a mechanism in which SOD1 is transported into lysosomes via autophagy. There, SOD1 helps clear lysosomal reactive oxygen species (ROS) and protects lysosomal function, suggesting a novel pathogenic mechanism for SOD1 mutation-induced ALS.
Their findings were recently published in the Journal of Cell Biology, entitled “SOD1 is delivered to lysosomes via autophagy to maintain lysosomal function and integrity.”
Neurodegenerative diseases arise from the degeneration of neuronal structure or function in the brain or spinal cord. A common hallmark in affected patients is the accumulation of protein aggregates in brain tissue, which is considered a major contributor to neurodegeneration. The lysosomal pathway is recognized as a key route for clearing these aggregates, yet the mechanisms for aggregate recognition and transport remain unclear.
To investigate the role of lysosomes in aggregate protein clearance, the research team generated reporters expressing aggregate proteins tagged with tandem mCherry-GFP. Given that the GFP signal is quenched in acidic environments, only the red mCherry signal is preserved if the substrate is delivered to lysosomes. Using this method, they discovered that disease-associated mutant SOD1 (G93A) enters lysosomes upon starvation. Surprisingly, wild-type SOD1 was also transported to lysosomes upon starvation (Figure 1), but unlike the mutant form, it was not significantly degraded once inside.
Further investigation revealed that SOD1 retains its superoxide dismutase activity under acidic conditions, and enzymatic activity was also detectable in isolated lysosomes (Figure 1). This suggests SOD1 may be transported to lysosomes to act as a superoxide dismutase and clear lysosomal ROS. Through lysosomal activity and ROS level assays, the researchers confirmed that SOD1 clears lysosomal ROS and protects lysosomal function from the ROS toxicity. This ROS clearance by SOD1 also helps protect lysosomes against membrane damage induced by LLoMe. Importantly, SOD1 mutations significantly reduced its activity under both neutral and acidic conditions, potentially representing a novel pathogenic mechanism of ALS.
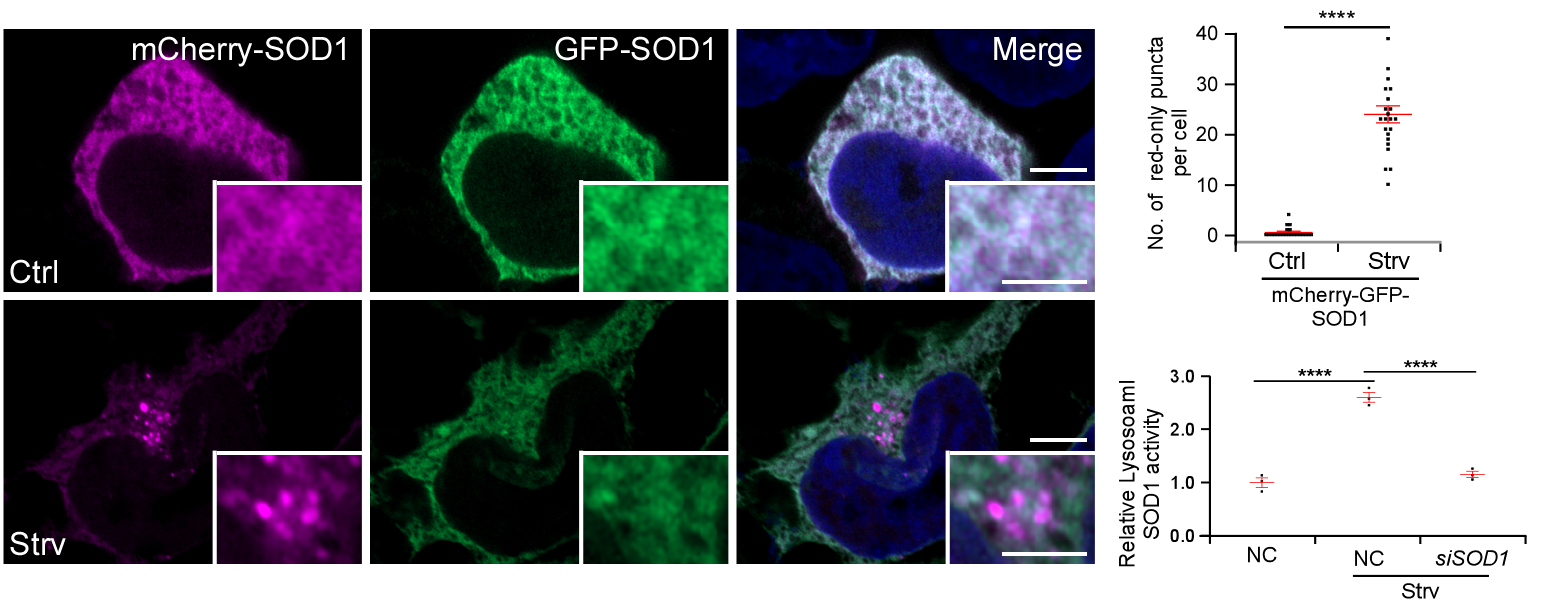
Figure 1. SOD1 enters lysosomes during starvation and retains superoxide dismutase activity
To further explore the mechanism of SOD1 transport into lysosomes, they performed a genome-wide CRISPRi screen. Screen results revealed that SOD1 is primarily delivered to lysosomes via the autophagy pathway. The selective autophagy receptor TP53INP1 mediates SOD1 recognition (Figure 2). TP53INP1 binds LC3 family proteins and SOD1 via its N-terminus and C-terminus, respectively, facilitating the targeting of cytosolic SOD1 to autophagosomes for subsequent lysosomal delivery. This process resembles the vacuolar delivery of hydrolases via autophagy in yeast, revealing a novel function for autophagy in maintaining lysosomal activity in mammalian cells.
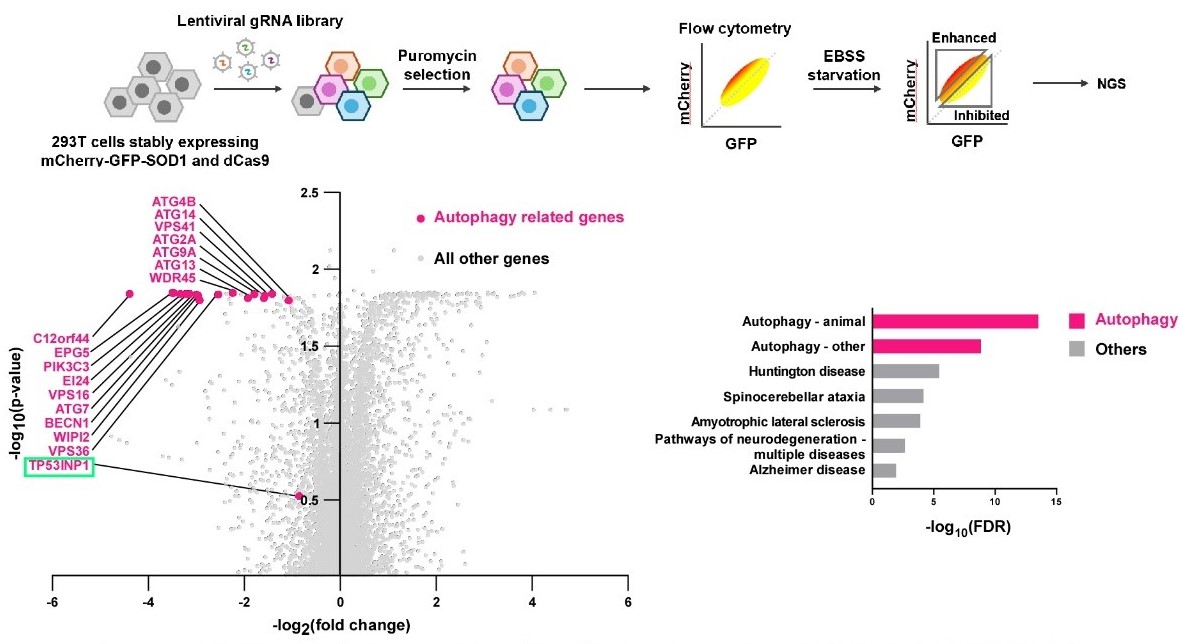
Figure 2. Genome-wide CRISPRi screen identifies that the autophagy pathway and specific receptor TP53INP1 are involved in SOD1 delivery to lysosomes
This work uncovers a pathway where SOD1, recognized by the specific receptor TP53INP1, enters lysosomes via autophagy to clear lysosomal ROS and protect lysosomal activity (Figure 3). This study not only advances our understanding of SOD1 function but also suggests potential mechanisms for SOD1 mutation-induced ALS and reveals a novel function of cellular autophagy.
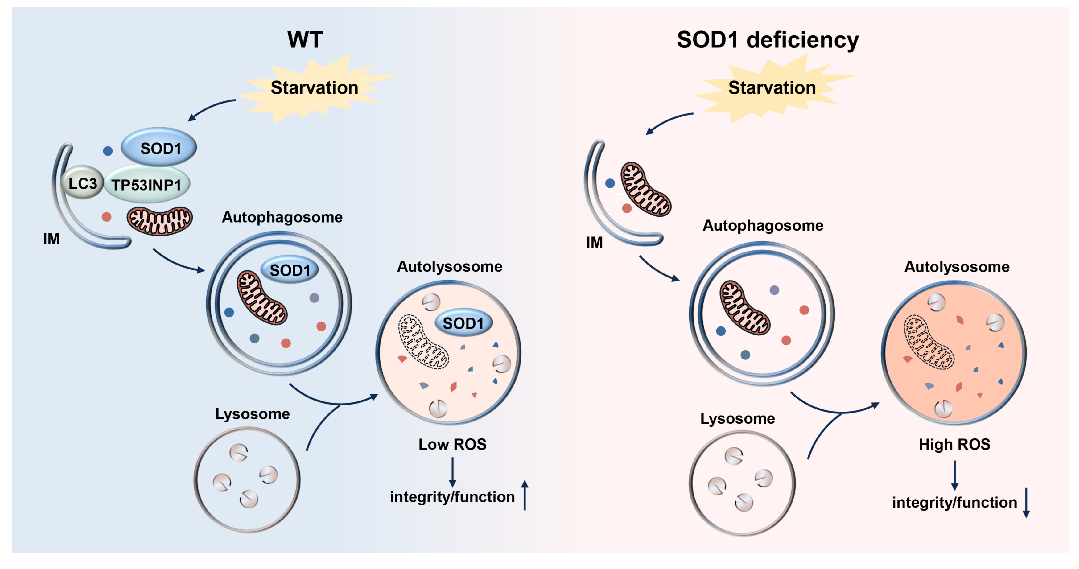
Figure 3. Schematic model of SOD1 transport into lysosomes during starvation and its function
Doctoral student Yanzhe Zheng is the first author of this paper, with Associate Professor Yan Zhao and Assistant Professor Ruilin Tian serving as the co-corresponding authors. Additional co-authors include postdoctoral researcher Meng Li, doctoral students Xuelin Chen and Ze Zheng, and undergraduate student Zixuan Chen, all from SUSTech.
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yifei REN
Photo ByYan QIU