Aqueous environmental nucleic acid (eNA), which refers to the DNA/RNA released into the water environment from surrounding species, contains a wealth of bioinformation. With advantages such as non-invasiveness, ease of sampling, and high information content, aqueous eNA-based technology has been developed and widely utilized for ecological research, pathogen surveillance, and pollution tracing, influencing management and policy decisions. At the same time, responsive environmental management demands a constant stream of information about the living world, which require large-scale, high-frequency sampling and testing of eNA, thus necessitating easy-to-operate, low-cost, and efficient eNA extraction techniques.
Current techniques face significant challenges due to the large sample volumes, low concentrations, and diverse states of eNAs in environmental water. Common nucleic acid extraction methods, such as ethanol precipitation, spin column, and traditional magnetic nanoparticles, are not suitable for extracting eNA from large volumes of environmental water samples due to their high cost and time-consuming procedures.
The most common approach is membrane filtration, in which intact cells from water samples are captured by 0.2 μm pore-sized membrane filter. However, a notable proportion of extracellular eNA (e-eNA) can pass through the membrane, remaining in the filtrate, such as cell-free nucleic acids and those encapsulated within viral particles. These e-eNA carry important biological information about microbial community structure and functional attributes such as antibiotic resistance gene (ARG). Their poor recovery limits applications in waterborne virus surveillance. Current methods available for e-eNA enrichment, including chemical precipitation, ultracentrifugation, and ultrafiltration, remain complex, expensive, and difficult to scale up.
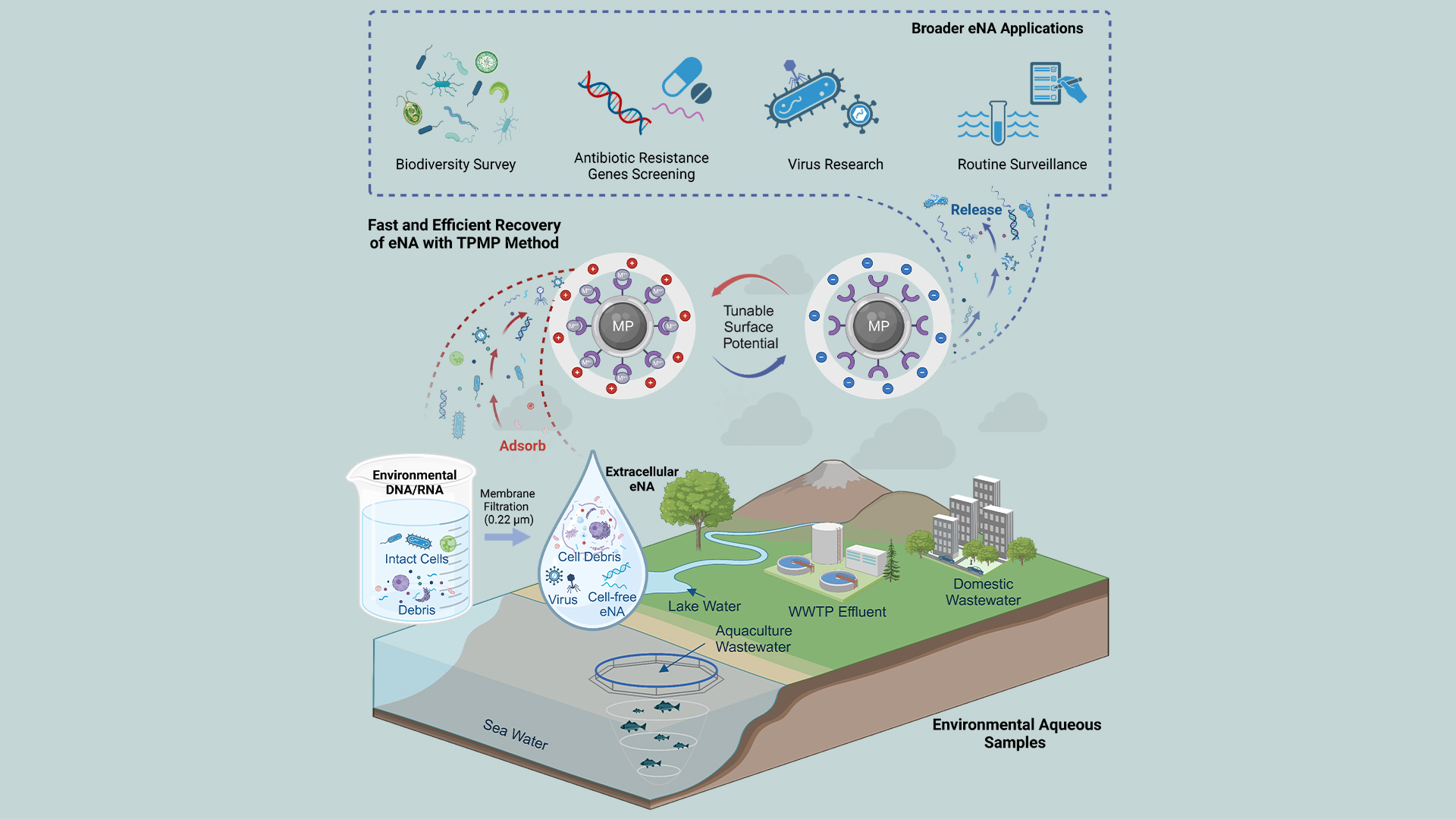
To address these limitations, Associate Professor Bo Zhang’s research team from the Department of Biomedical Engineering at the Southern University of Science and Technology (SUSTech) has developed an extraction method based on tunable surface potential magnetic nanoparticles (TPMPs). This approach achieves approximately 90% recovery efficiency within 60 minutes, with superior enrichment across various eNA forms, including cell-free DNA/RNA, intracellular nucleic acids, and viral nucleic acids. Fast, efficient, reusable, and low-cost, the TPMP-based technique holds significant potential for applications in environmental nucleic acid surveillance.
Their work, entitled “Tunable Surface Potential Nanomaterials for Enhanced Environmental Nucleic Acid Extraction and Surveillance,” has been published in the renowned international journal Water Research.
The researchers developed an aqueous eNA extraction method based on TPMP, achieving up to a 90% recovery rate and 100- to 1000-fold enrichment of eNAs. The TPMP method demonstrated high recovery efficiency for various forms of eNA, including cell-free DNA/RNA with different fragment lengths, intracellular eNA, and virion-encapsidated eNA. Applying this technique, they extracted environmental DNA (eDNA) from five different types of real-world aqueous samples, which demonstrated a higher eDNA yield—improving by 1.2 to 11.8 times—compared to commonly used methods, while also reducing the extraction time from several hours to less than 60 minutes. Importantly, TPMP was successfully recycled and reused, maintaining consistent performance across multiple extraction cycles.
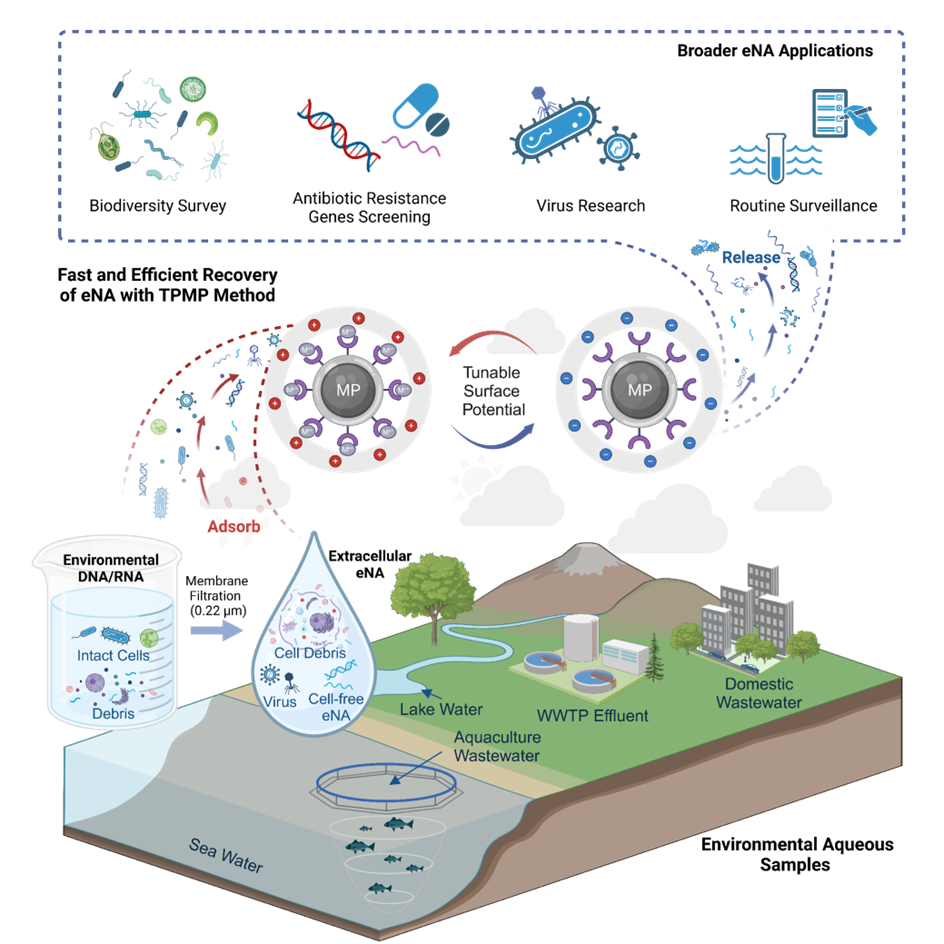
Figure 1. Environmental DNA/RNA (eNA) and extracellular eNA extracted from diverse water samples using the TPMP method, applied across various scenarios
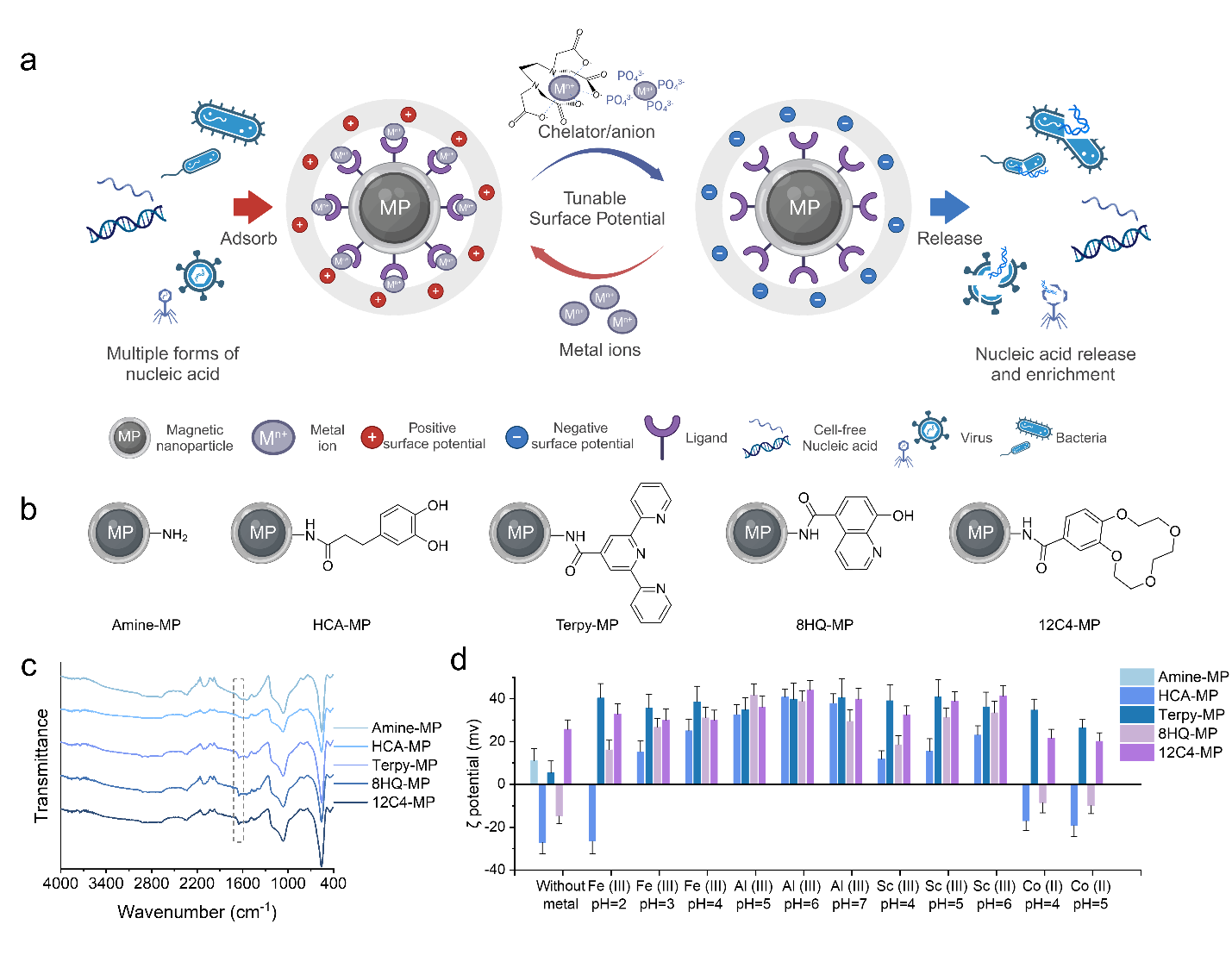
Figure 2. Development and characterization of TPMP
Beyond improving recovery rates, the TPMP method’s also enabled deeper investigation of extracellular eNAs, which are often overlooked in traditional studies. The results revealed that e-eNA not only harbors a distinct microbial composition compared to intracellular eNA (i-eNA), providing bioinformation from lysed microbes under increased environmental pressure. Moreover, e-eNAs carry genetic information from lysed hosts and associated viruses, offering valuable data for comprehensive virus-host interaction analyses. Additionally, high-risk ARG, particularly those linked to viruses and pathogens, were found to be concentrated in e-eNAs, underscoring the importance of e-eNA monitoring.
Leveraging the TPMP method’s cost-efficiency, time-saving operation, and ease of use, the team established a routine environmental monitoring platform for long-term ARG detection. The platform successfully identified both baseline levels and seasonal fluctuations of ARG hotspots. These findings indicate that the TPMP approach meets the growing demand for eNA analysis and expands its applications in ecological surveys, e-eNA research, ARG and pathogen screening, and routine environmental monitoring.
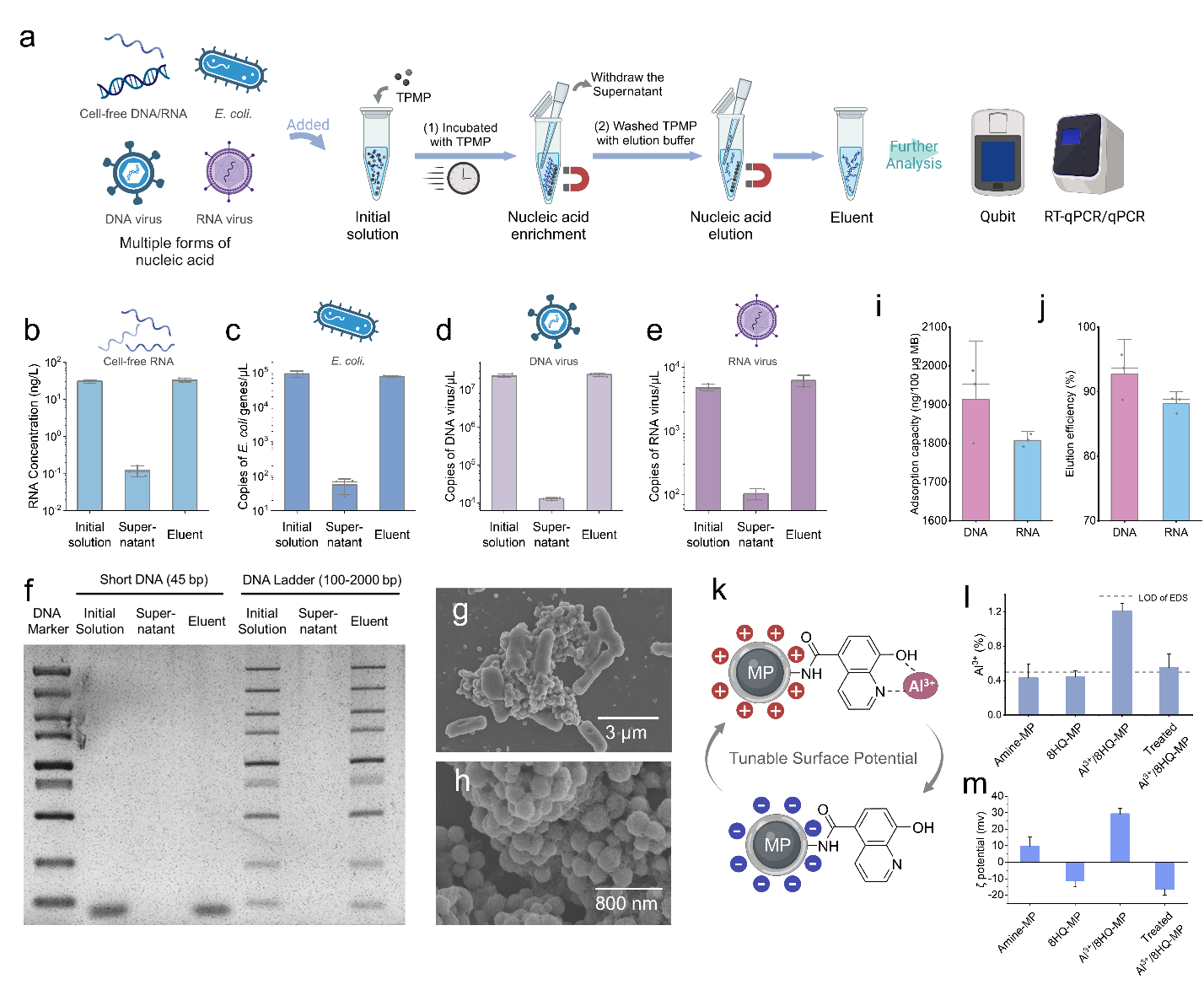
Figure 3. Performance of the TPMP-based nucleic acid extraction method
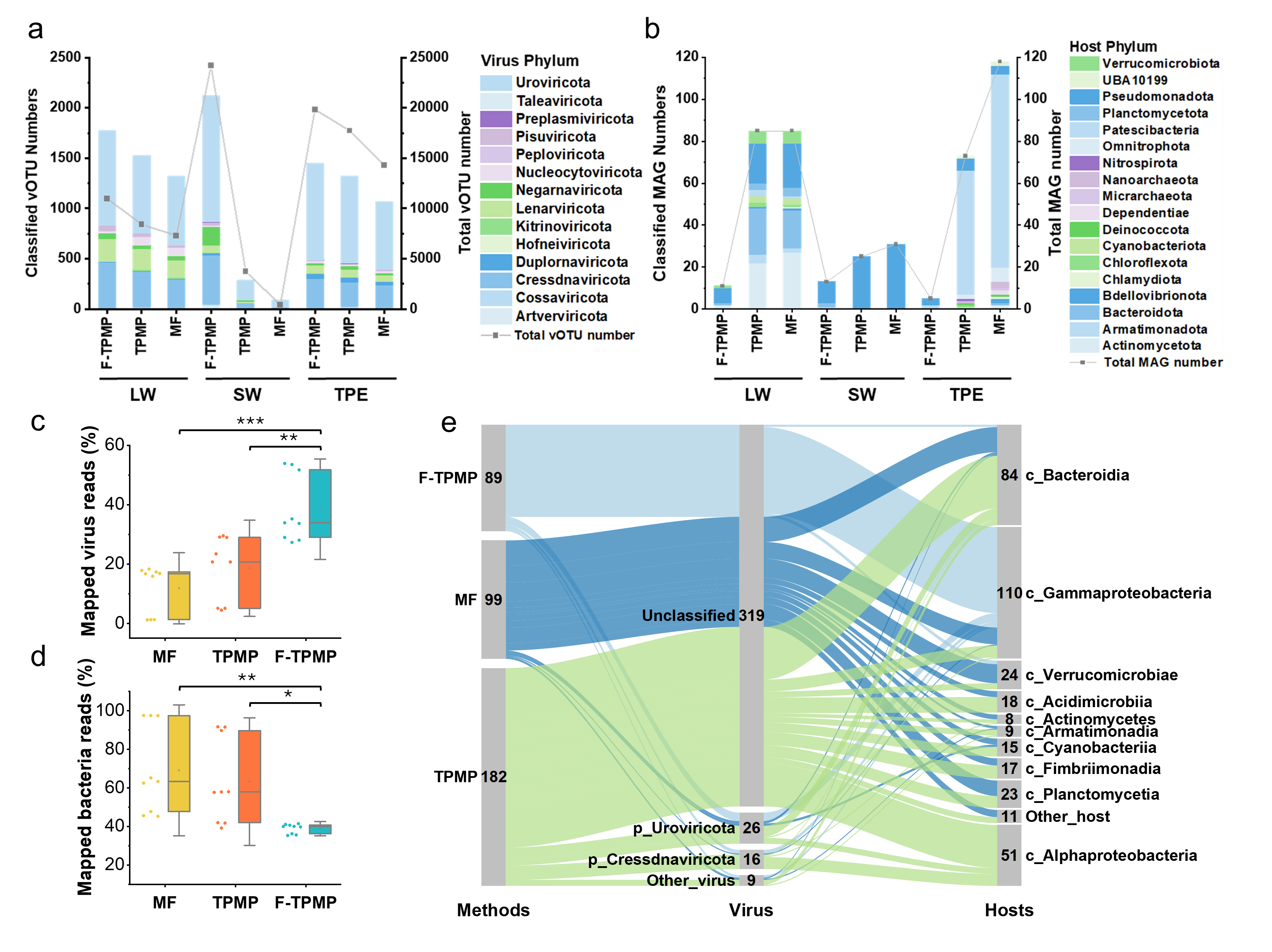
Figure 4. Virus-host analysis based on extracellular and intracellular eDNA
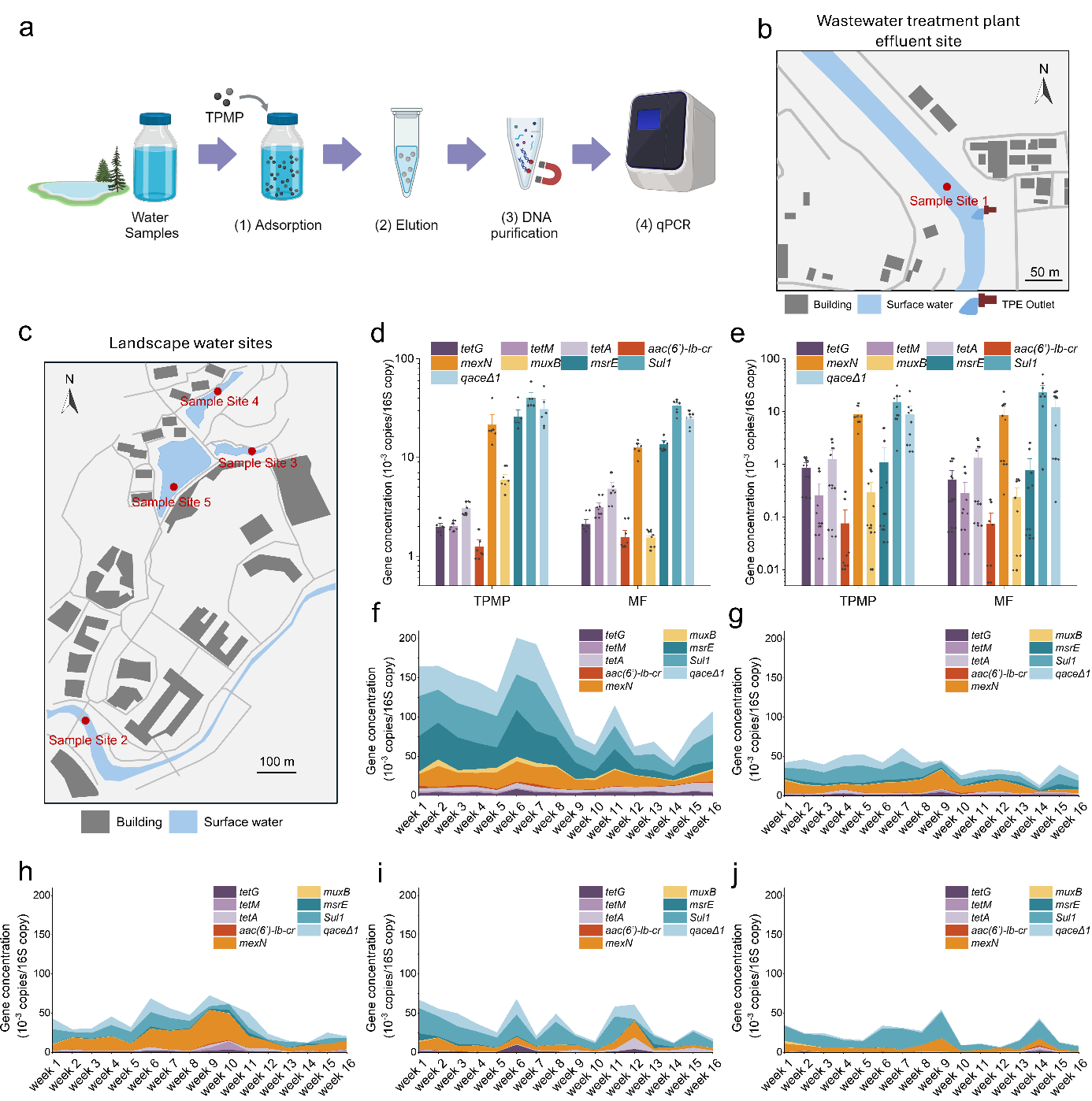
Figure 5. Routine monitoring of ARG with TPMP method
Doctoral student Jiahui Lv, master’s student Tong Li, and Research Assistant Professor Ying Liu from the Department of Biomedical Engineering at SUSTech are the co-first authors of this paper. Associate Professor Bo Zhang is the corresponding author, while Professor Pan-Lin Shao from Guangzhou Medical University (GMU) is the co-corresponding author. SUSTech is the primary affiliated institution for this work.
Paper link: https://doi.org/10.1016/j.watres.2025.124428
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yuwen ZENG
Photo ByDepartment of Biomedical Engineering