Recently, Dr. Xin Gong’s group from the Department of Biology at the Southern University of Science and Technology (SUSTech) published a research article entitled “Structural basis of substrate recognition and translocation by human ABCA4” in Nature Communications. They’ve presented three cryo-EM structures of human ABCA4, a retina-specific ABCA transporter, in distinct functional states at atomic resolutions. The study provides a molecular basis to understand the mechanism of ABCA4-mediated substrate recognition and translocation and suggests a common “lateral access and extrusion” mechanism for ABCA-mediated lipid transport.
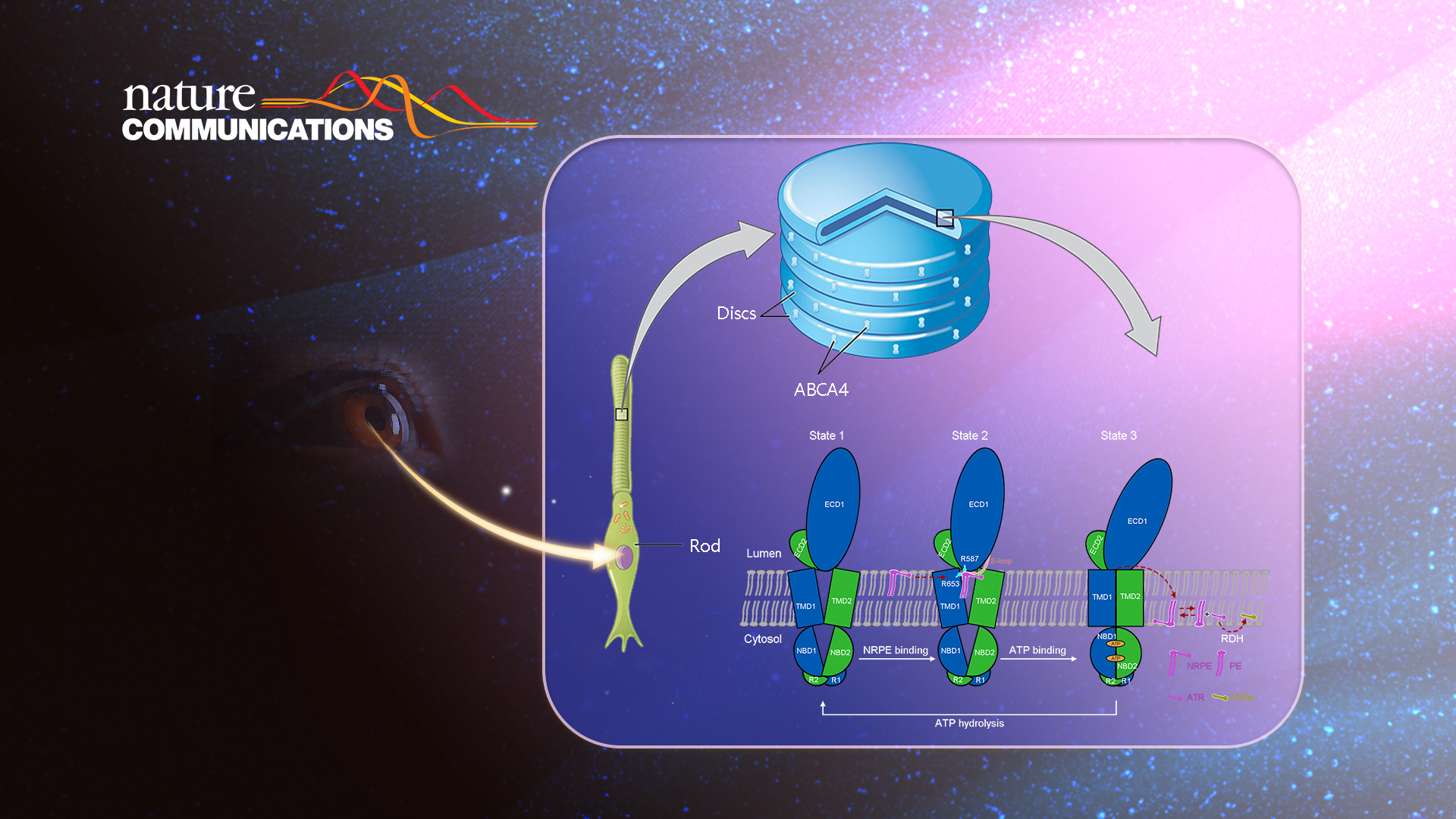

ABCA4 (also known as ABCR or the rim protein) is a retinal-specific member of the ABCA subfamily predominantly expressed in photoreceptor cells’ outer segment disk membranes. The ABCA4 gene is related to Stargardt disease, the most common heritable macular degenerative disorder, and implicated in several other severe visual disorders, such as age-related macular degeneration, retinitis pigmentosa, and cone-rod dystrophy.
Available evidence suggested that ABCA4 may act as a flippase to translocate N-retinylidene-phosphatidylethanolamine (NRPE), a reversible covalent adduct of all-trans-retinal (ATR) and phosphatidylethanolamine (PE), from the luminal leaflet to the cytoplasmic leaflet of the disk membrane. After translocation, NRPE can be hydrolyzed to ATR and PE, thereby enabling ATR to be reduced to all-trans-retinol by the cytoplasmic retinal dehydrogenase.
Through this proposed model, the toxic ATR in photoreceptor cells following photoexcitation is removed from the luminal leaflet of the disc membrane and then goes into the retinoid cycle for regeneration of 11-cis-retinal. Otherwise, ATR and NRPE would accumulate in the luminal membrane leaflet and form toxic bisretinoid derivatives that may cause retinal degeneration. However, the molecular basis for substrate recognition by ABCA4 requires further investigation.
The researchers obtained the human ABCA4 protein by recombinant expression and purification. The in vitro ATPase activity of ABCA4 and the corresponding enzyme kinetic parameters were measured in the study. Then the researchers used single-particle cryo-EM to determine the high-resolution three-dimensional structures of human ABCA4 in apo, NRPE-bound, and ATP-bound states (Figure 1).
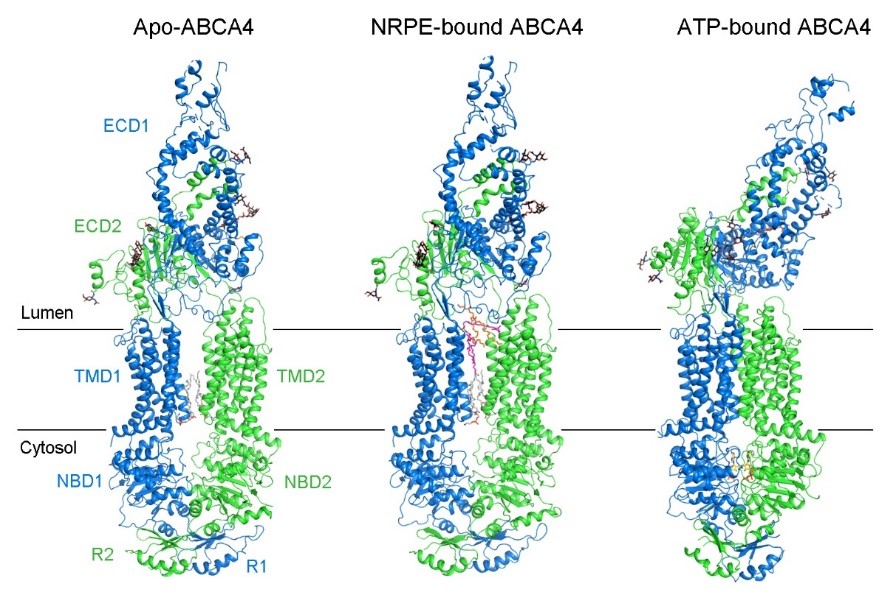
Figure 1. The structure of human ABCA4 in apo, NRPE-bound, and ATP-bound states
To reveal the molecular basis for substrate recognition, the researchers further verified the importance of the substrate-binding sites by ATR-stimulated ATPase activity of ABCA4 (Figure 2). The NRPE-bound ABCA4 structure shows that both NRPE and PL3 molecules are sandwiched between the two TMDs in the luminal leaflet, and completely exposed to the lipid bilayer, suggesting the lateral access of the lipid substrates from the luminal membrane leaflet (Figure 2b).
The phosphate group of NRPE is mainly recognized by the side chains of the positively charged Arg587 and Arg653 residues, whereas the acyl chains are coordinated by TMs 1/2/5 via extensive hydrophobic interactions (Figure 2c, left). The ATR moiety of NRPE interacts with the four aromatic residues on the S-loop (Trp339, Tyr340, Tyr345, and Phe348) and the hydrophobic residues in TM8 and TM11 (Fig. 2c, right). In the presence of PE, ATR can stimulate the ATPase activity of ABCA4 (Figure 2d).
To reveal the function of the NRPE binding sites in ABCA4, the researchers generated 6 ABCA4 variants with point mutations designed to disturb NRPE binding, including R587A, R653C, and R587A/R653C designed to disrupt the polar interactions with the phosphate group of NRPE; W339E/Y340E, Y345E/F348E, and S1677E/I1812E designed to explore the hydrophobic interactions with the ATR moiety of NRPE. Although these variants retained certain basal ATPase activity, they displayed nearly abolished ATR-stimulated ATPase activity (Figure 2d).
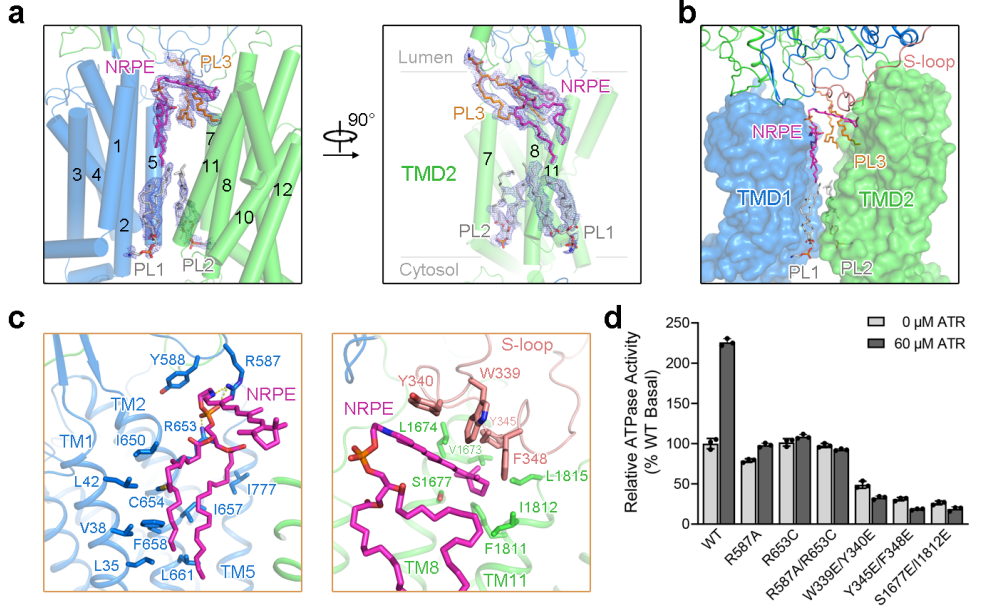
Figure 2. NRPE substrate binding site
The researchers further proposed a model of NRPE translocation by ABCA4 based on the structural analyses of ABCA4 in three different functional states (Figure 3). In the apo state, the TMDs exhibit a V-shaped lateral-opening conformation, allowing the lateral entry of NRPE from the luminal membrane leaflet (state 1). NRPE is recognized through two positively charged Arg residues in TMD1 and ECD1 (Arg653 and Arg587) and coordinated by two TMDs and S-loop of ECD1 through extensive hydrophobic interactions (state 2). Upon binding ATP, the rearrangement of TMDs accompanied with ATP-induced NBD closure transmits the TMDs to a closed conformation, which precludes the NRPE binding (state 3). The research provides a structural basis for the functional study of ABCA4 and related drug development and gives insight into the substrate recognition and translocation mechanism of the ABCA subfamily transporters.
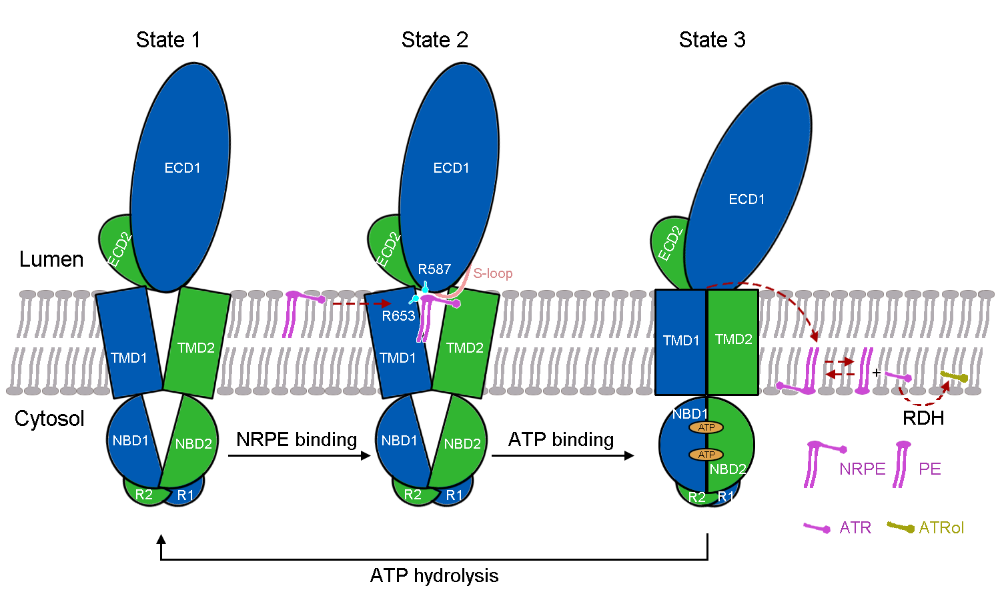
Figure 3. A working model of ABCA4-mediated NRPE translocation
This research was undertaken by the Department of Biology at SUSTech. Professor Xin Gong is the only corresponding author of the paper. Research Associate Professor Tian Xie and postgraduate student Zike Zhang are the co-first authors of this paper. This work is supported by the National Natural Science Foundation of China (NSFC), the Natural Science Foundation of Guangdong Province of China for Distinguished Young Scientists, and the Shenzhen Science and Technology Program. Cryo-EM data collection and image processing were supported by the Cryo-Electron Microscopy Center of SUSTech.
Paper link: https://www.nature.com/articles/s41467-021-24194-6.
Proofread ByAdrian Cremin, Yingying XIA
Photo By