Transcription and its associated RNA processing are key events during the decoding of genetic information in eukaryotes, which eventually links genotype to phenotype. In plants, it is well known that transcription factors regulate various biological processes by modulating transcription initiation. However, the regulation of transcription elongation and termination, both in terms of mechanisms and biological significance, remains unclear in plants. Furthermore, RNA processing is tightly coupled with transcription in both animals and plants, with a typical example being co-transcriptional splicing, where transcription and RNA splicing occur simultaneously.
Co-transcriptional splicing not only removes introns but also delivers protein to mark the spliced RNAs, thereby further regulating the fate of RNA after transcription, such as degradation and nuclear export. Plant genes typically have short introns, with an average length of only one-tenth that of mammals. Interestingly, co-transcriptional splicing occurs effectively in plants as well, but the underlying mechanisms are unclear. In addition, unlike mammals, plant Pol II often pauses (3′ end pausing) within a narrow region (250bp) downstream of the polyA site. Previously, such pausing was generally considered as a simple reflection of efficient transcription termination in plants. To date, it is still unclear what mechanisms are involved in 3′ pausing, how it is regulated, and how it relates to the transcription termination process.
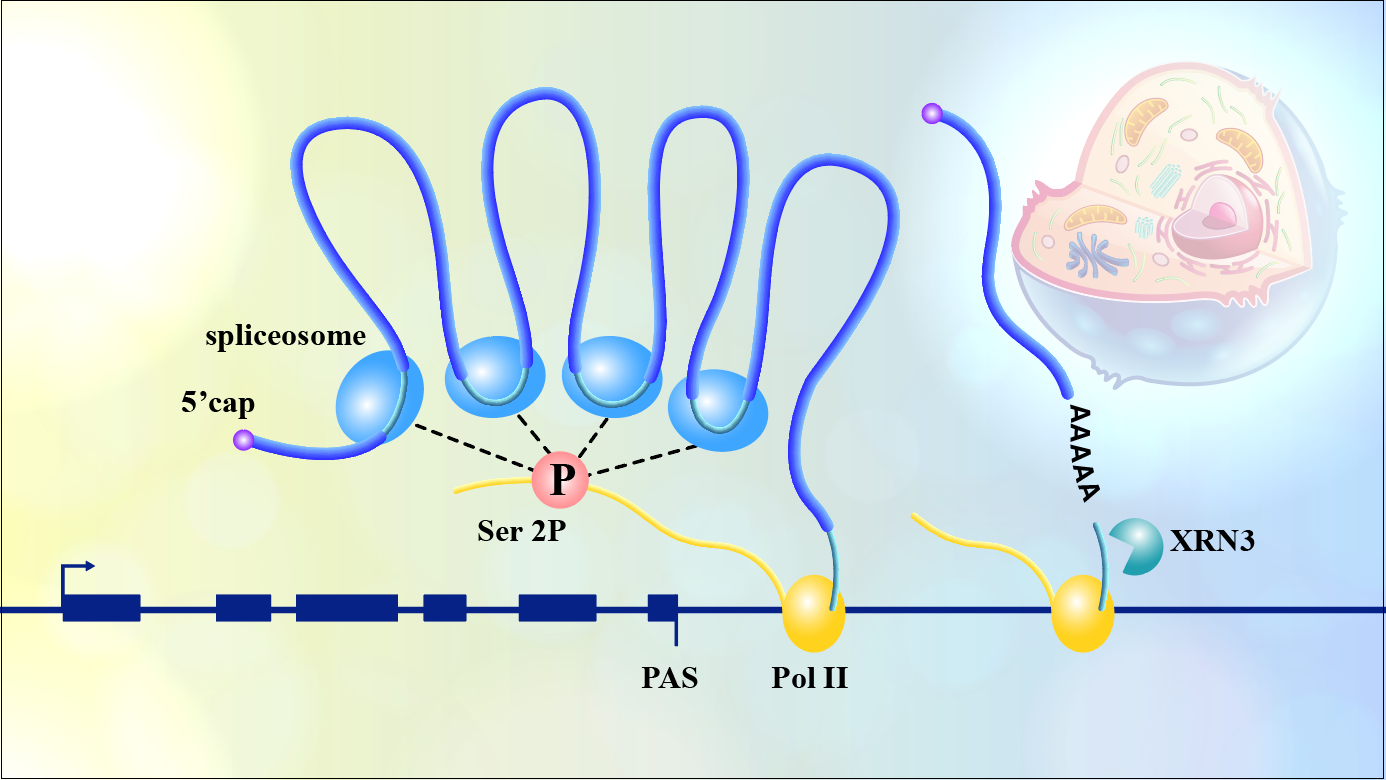
Associate Professor Zhe Wu’s research group from the School of Life Sciences at the Southern University of Science and Technology (SUSTech), in collaboration with Assistant Professor Ziwei Dai’s research group from the Department of Biology, recently combined comprehensive experiments with mathematical modeling to explore the characteristics and regulatory mechanisms of the pausing occurring at the 3′ end of genes during the transcription termination stage of the model plant Arabidopsis. This research revealed that Pol II pausing at the 3′ end of genes is a crucial checkpoint for ensuring efficient co-transcriptional splicing of multi-intron genes in plants.
Their research paper, entitled “Coupling of co-transcriptional splicing and 3′ end Pol II pausing during termination in Arabidopsis”, has been published in the internationally renowned academic journal Genome Biology.
The study first utilizes the single-nucleotide resolution pNET-seq (Plant Native Elongating Transcripts Sequencing) technology to obtain a high-precision map of Pol II distribution at the 3′ end of genes. It uncovers that Pol II pausing at the 3′ end of genes is an important feature of transcription in Arabidopsis, and defines the strength of Pol II pausing at the 3′ end using the 3′ Pause Index (3′ PI). The analysis reveals that the position rather than the strength of Pol II pausing at the 3′ end of genes is related to transcription termination efficiency. Furthermore, in the transcription termination-defective mutant xrn3, Pol II pausing at the 3′ end of genes was not strongly affected, indicating that although Pol II pausing at the 3′ end of genes is closely associated with transcription termination, there are still other factors regulating the strength of Pol II pausing at the 3′ end of genes.
Further analysis revealed a strong correlation between the degree of Pol II pausing at the 3′ end of genes and the number of exons present in the genes (Figure 1a, b, c). Genes with a higher number of exons exhibit a higher degree of Pol II pausing at the 3′ end. Interestingly, Prof. Wu’s research group had previously reported that the number of exons in a gene is closely associated with the efficiency of co-transcriptional splicing, with genes having more exons tending to have higher co-transcriptional splicing efficiency. Further analysis in this study reveals that there is a positive correlation between gene exon numbers, co-transcriptional splicing efficiency, and 3’ end Pol II pausing levels (Figure 1d), suggesting that the number of exons in a gene and co-transcriptional splicing efficiency may be important factors influencing Pol II pausing at the 3′ end of genes.
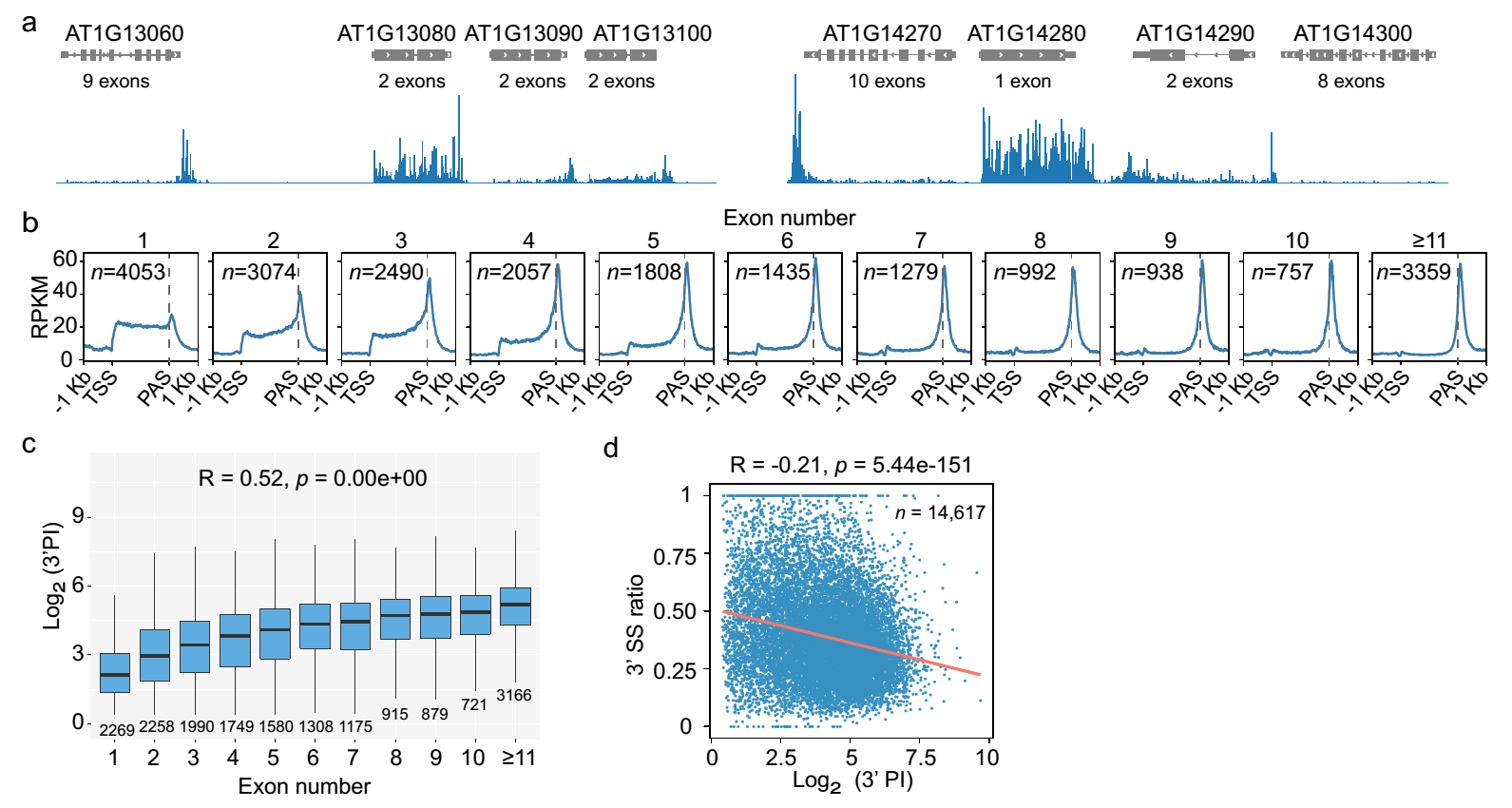
Figure 1. 3’end Pol II pausing correlates gene exon numbers and co-transcriptional splicing efficiency
The researchers used herboxidiene to treat plants, which inhibits the assembly of the spliceosome and thus inhibits splicing. It was found that after treatment with herboxidiene, the level of Pol II pausing at the 3′ end of genes dramatically decreased. Additionally, the correlation between the number of exons in genes and the strength of Pol II pausing at the 3′ end disappeared (Figure 2), indicating that the efficient assembly of the spliceosome is necessary for Pol II pausing at the 3′ end of genes.
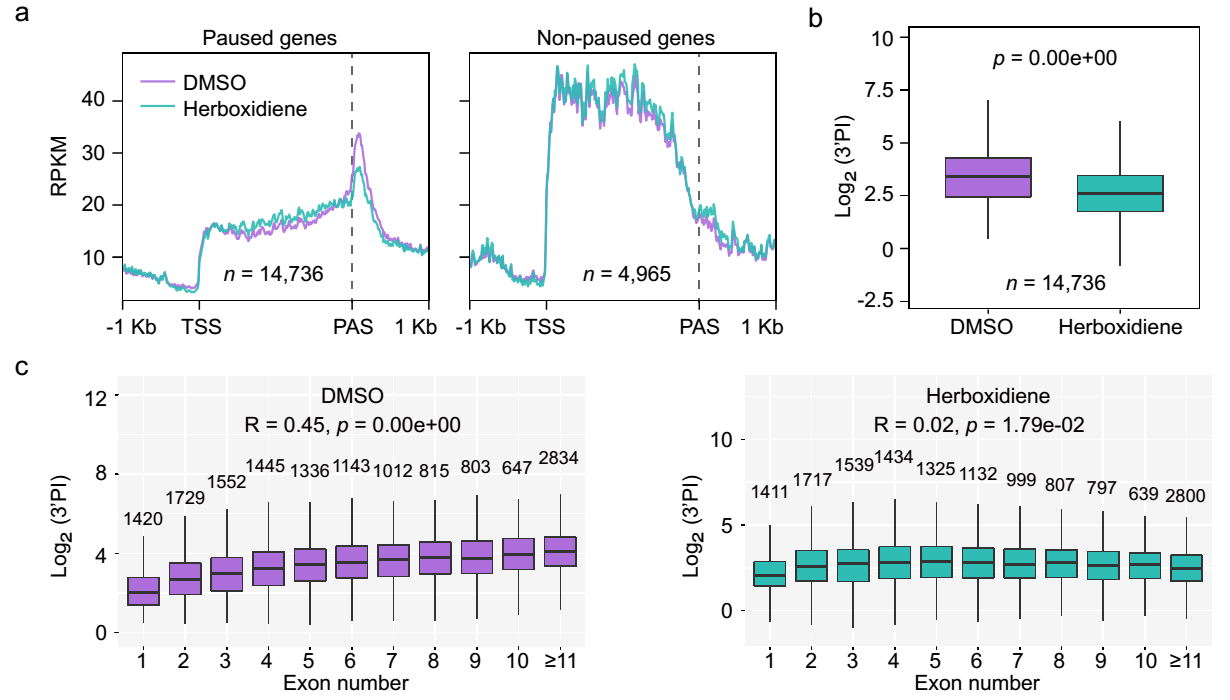
Figure 2. Treatment of a splicing inhibitor, herboxidiene, reduces the level of 3’end Pol II pausing and disrupts its correlation with gene exon numbers
Finally, the study built a mathematical model that is able to simulate the process of Pol II transcription elongation and termination. The model was used to fit the experimental data of Pol II distribution, and the results of the fitting further determined various parameters of Pol II transcription in different conditions and genotypes. The fitting results further supported the conclusion that the number of exons is an important factor influencing the duration of Pol II pausing at the 3′ end of genes.
In conclusion, this work proposes that the number of exons in a gene, coupled with co-transcriptional splicing, determines the strength of Pol II pausing at the 3′ end of genes. Effective assembly of the spliceosome is necessary for Pol II pausing at the 3′ end of genes. Pol II pausing at the 3′ end of genes may serve as a checkpoint for efficient co-transcriptional splicing at multi-exon genes (Figure 3). The research has elucidated the biological significance and regulatory mechanisms of Pol II pausing at the 3′ end of genes in plants.
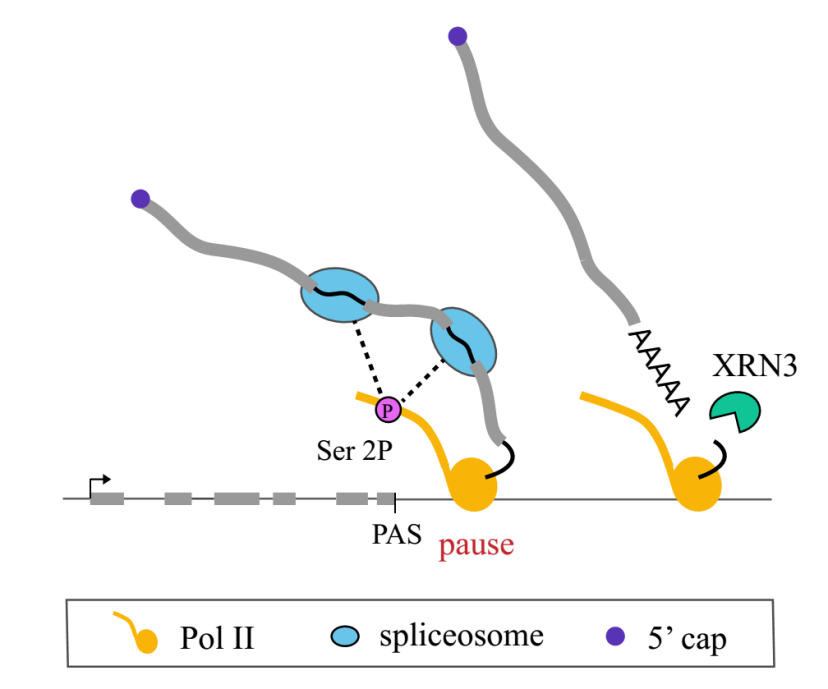
Figure 3. A working model of 3’ end Pol II pausing in Arabidopsis
Ph.D. student Sixian Zhou and postdoctoral scientist Fengli Zhao (now an Assistant Researcher at the China National Rice Research Institute) from Prof. Zhe Wu’s research group at SUSTech are the co-first authors of this paper. Profs. Zhe Wu’s and Ziwei Dai’s groups performed the experiments and mathematical modeling, respectively, and are the co-corresponding authors. Research Associate Professor Danling Zhu and Ph.D. student Qiqi Zhang from SUSTech also contributed to this research.
This work was supported by the National Natural Science Foundation of China (NSFC), Guangdong Innovation Research Team Fund, Shenzhen Innovation Committee of Science and Technology, and the Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes.
Paper link: https://doi.org/10.1186/s13059-023-03050-4
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By