Eukaryotic cells encapsulate their genetic information within chromatin, forming a ‘beads-on-a-string’ arrangement known as nucleosomes. Each nucleosome consists of 147 bp DNA wrapped around the histone octamers (H2A, H2B, H3, and H4)2. Post-translational modifications of histones in nucleosomes are vital for regulating chromatin structure and gene expression, significantly impacting the development and progression of diverse diseases.
SUV420H1, a member of the SUV4-20H family protein, is primarily responsible for catalyzing the bulk of H4K20me2 and H4K20me3 modifications. Recent research has shown that the histone variant H2A.Z enhances SUV420H1’s catalytic activities, particularly in the deposition of the H4K20me2 mark. This enhancement is crucial for the recruitment of ORC1, thereby regulating DNA replication. However, the specific mechanism through which SUV420H1 recognizes and methylates H4K20 at the nucleosome level remains largely elusive. Similarly, the enhancement of SUV420H1 activities by H2A.Z-containing nucleosomes is still not fully understood.
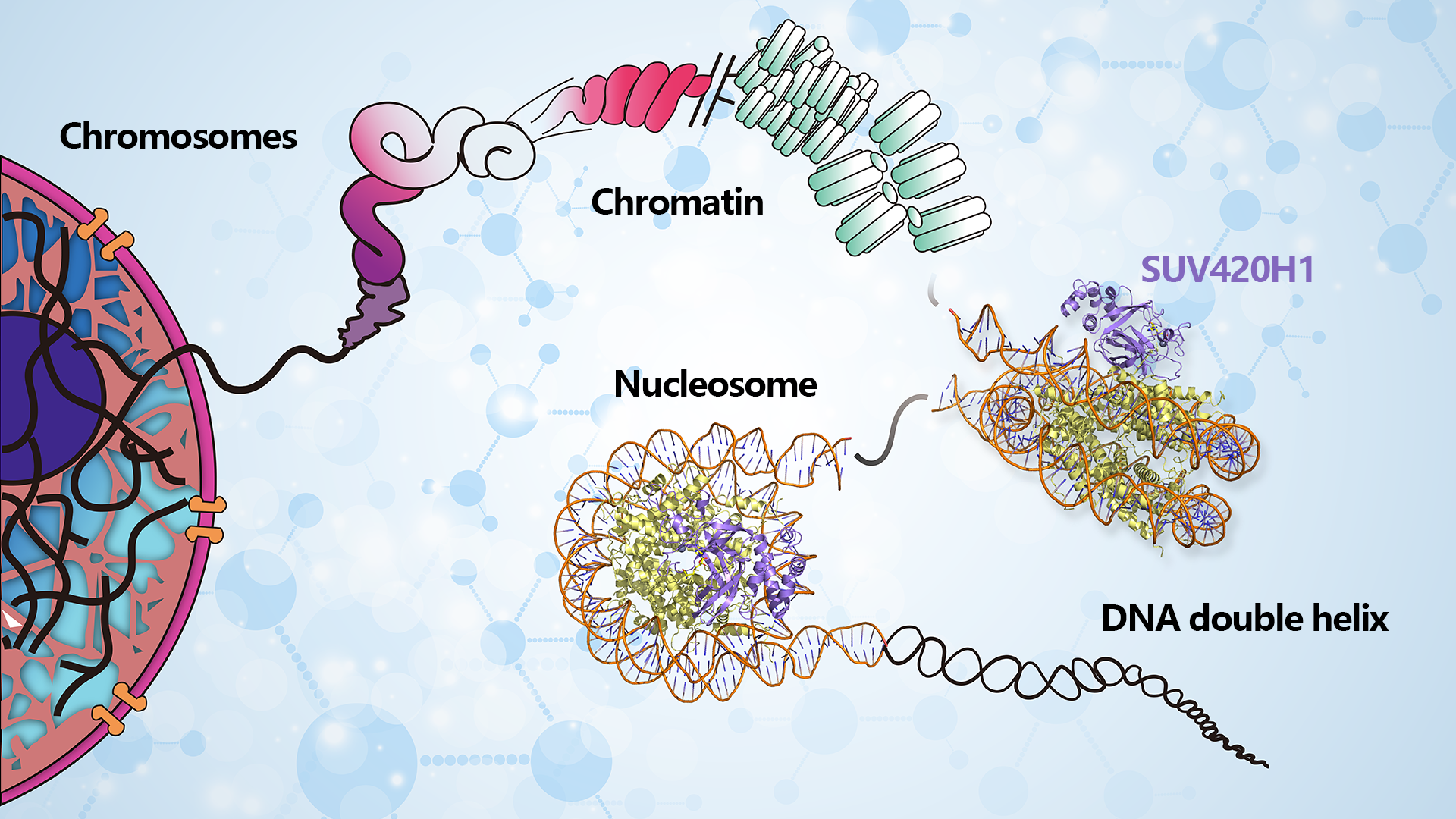
Assistant Professor Wanqiu Li from the School of Medicine at the Southern University of Science and Technology (SUSTech), in collaboration with Associate Professor Zengqi Wen from Sun Yat-sen University’s Medical School, has recently published their latest research that unveils high-resolution cryo-EM structures of the SUV420H1 respectively complexed with classic or H2A.Z-containing nucleosomes. The results illuminate the nucleosome-based recognition and methylation mechanisms of SUV420H1 and how the H2A.Z nucleosome enhances SUV420H1’s binding and catalytic activity, laying a molecular foundation for better understanding diseases related to SUV420H1.
Their research work, entitled “Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase”, has been published in Cell Discovery.

Figure 1. The overall structure of SUV420H1 in complex with H2A.Z-containing nucleosomes. (a) Cryo-EM density map and (b) atomic model.
Their research analyzed the cryo-EM structures of the SUV420H1 in complex with either H2A-containing or H2A.Z-containing nucleosomes (Figure 1). In these structures, SUV420H1 is observed interacting with the nucleosomal DNA, the N-terminal tail of H4, and the acidic patch of H2A/H2A.Z-H2B. Specifically, the residue R286 of SUV420H1 engages with the nucleosomal DNA. The arginine anchor residues, R352 and R357, in SUV420H1, are specifically targeted towards the acidic patch of H2A/H2A.Z-H2B. Furthermore, S255 and S283 are positioned at the entrance of that H4 tail insertion into the catalytic pocket.
Mutations S255F and S283L, which are associated with breast and lung cancers, have larger side chains that block the entry of the H4 tail into the catalytic pocket of SUV420H1, thereby impairing its methylation activity (Figure 2a and 2d). Drawing from these structural insights, a series of mutations were designed to explore their functions, revealing various degrees of reduction in the catalytic activity and binding capacity of SUV420H1 (Figure 2b and 2c).

Figure 2. The interaction interface of SUV420H1 with nucleosomes and the validation of its function in vivo and in vitro.
Notably, comparative analysis of the SUV420H1-H2A and SUV420H1-H2A.Z nucleosome complexes revealed a subtle conformational change in the amino acid residue R220. In the SUV420H1-H2A.Z nucleosome complex, the distance between R220 and E94 is closer compared to that between R220 and E91 in the SUV420H1-H2A nucleosome complex. This observation was further substantiated by in vitro biochemical assays and cellular immunoprecipitation experiments, which underscored the pivotal role of R220 in enhancing the catalytic activity of SUV420H1 on H2A.Z-containing nucleosomes. Moreover, ATAC-seq experiments revealed that certain mutations related to diminishing the catalytic activity of SUV420H1, including the R220A mutation, also impaired the activity of SUV420H1 in restoring the chromatin states (Figure 2).
Overall, the study elucidates the structural basis of SUV420H1’s recognition of classic and H2A.Z-containing nucleosomes and its catalysis of H4K20 methylation modifications. It also identifies the interaction interface and key amino acid residues that promote the enhanced catalytic activity of H2A.Z-containing nucleosomes on SUV420H1. This provides a solid foundation for further research on the function of SUV420H1 and its role in chromatin regulation.
Ph.D. student Folan Lin from the School of Medicine at SUSTech is the first author of this paper. Assistant Professor Wanqiu Li and Associate Professor Zengqi Wen are co-corresponding authors. Ruxin Zhang, a joint doctoral student from Shenzhen Bay Laboratory and Capital Medical University, and Weihan Shao, Cong Lei, Mingxi Ma, and Ying Zhang, all from the School of Medicine at SUSTech, contributed to this study. SUSTech is the primary institution for this research, and collaborating institutions include Sun Yat-sen University.
This work was supported by the National Natural Science Foundation of China (NSFC) and the Science, Technology and Innovation Commission of Shenzhen Municipality. The cryo-electron microscopy data collection was supported by the Cryo-Electron Microscopy Center at SUSTech.
Paper link: https://www.nature.com/articles/s41421-023-00620-5
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo BySchool of Medicine