Raman spectroscopy detects the molecular bond information of the chemical components of the sample in situ in a non-destructive and label-free manner. It is an emerging metabolic-related spectromics technology in biological and clinical medical research and is expected to promote the evolution of precision medicine. However, it is susceptible to interference from instruments, environmental noise, and background signals (non-Raman signals/baselines) due to weak Raman scattering signals.
The complexity of the components of biological samples and the severe interference of fluorescence signals make the biomedical application of Raman spectroscopy difficult. Spectral preprocessing methods with efficient spectral noise removal and high-fidelity baseline correction capabilities are the prerequisites and challenges for high-quality Raman spectroscopy applications.

Chair Professor Perry Ping Shum’s team from the Department of Electronic and Electrical Engineering (EEE) and the State Key Laboratory of Optical Fiber and Cable Manufacture Technology at the Southern University of Science and Technology (SUSTech) has recently made significant breakthroughs in general analysis algorithms for Raman spectroscopy. The team and their collaborators have proposed a two-step Raman spectral preprocessing strategy (RSPSSL) based on self-supervised learning. This method achieves high-fidelity spectral denoising and baseline correction across various instruments, samples, and spectral types, promoting the chemical resolution visualization of Raman hyperspectral images of clinical tissue samples.
Their article, “RSPSSL: A Novel High-fidelity Raman Spectral Preprocessing Scheme to Enhance Biomedical Applications and Chemical Resolution Visualization”, has been published in Light: Science & Applications. Additionally, their work was selected as the back cover of the February edition of the journal.

Back cover of the February edition of Light: Science & Applications
The first step of the scheme is to establish a self-supervised model that self-decomposes, rearranges, and reconstructs the unlabeled training spectra based on the mutual independence of the physical relationship between Raman peaks, noise, and baselines. This model uses a generative adversarial network to generate an infinite number of labeled high-simulation Raman spectral pairs, addressing the problem of unlabeled real Raman spectra. The label-free training spectra employs diverse data from multiple laboratories across instruments, samples, and spectral types to obtain the diversity of noise and baselines.
Secondly, to adapt to the complexity of actual spectral data, the preprocessing model enhances the fitting ability of complex signals through the end-to-end connection of multiple submodules. The preprocessing model RSBPCNN# can be used for Raman spectral preprocessing from any instrument, sample, and spectral type without manual intervention or retraining.

Figure 1. Overall framework diagram of the method proposed in this study
In this study, the researchers propose RSPSSL, a new strategy for self-supervised two-step Raman spectral preprocessing. This method generates an infinite number of labeled high-fidelity simulation spectral datasets through the fine separation and reconstruction of diverse spectral features. It trains and optimizes a predominant preprocessing model (RSBPCNN#) with high fitting ability that is capable of high-throughput of arbitrary Raman spectroscopy noise elimination and baseline correction without human intervention.
This high-fidelity approach significantly improves the accuracy of cancer diagnosis and solution concentration prediction in experiments, and improves the full-spectrum quality of hyperspectral images. It also eliminates the background signal of the biological silent zone, realizes the visualization of chemical resolution of images in the spectral fingerprint region, and reflects the broad-spectrum applicability across various instruments, samples, and spectral types.
In the future, incorporating the spatial distribution of spectra can further enhance the resolution of hyperspectral images. This method has been integrated into the laboratory-sharing platform for scientific use, allowing researchers to load Raman spectral data in batches to achieve rapid spectral preprocessing (1900 spectra/sec). For more information, see the related link below.
“This research has demonstrated approximately a tenfold performance improvement over existing Raman spectroscopy preprocessing algorithms. It enables broad-spectrum applicability across diverse instruments, samples, and spectral types, as well as ultrahigh chemical resolution visualization in biological tissue samples through multi-channel (Raman shift) analysis. This advancement will facilitate the clinical application and basic medical research of label-free Raman spectroscopy imaging, contributing to the transformation of precision medicine,” stated Professor Perry Ping Shum.
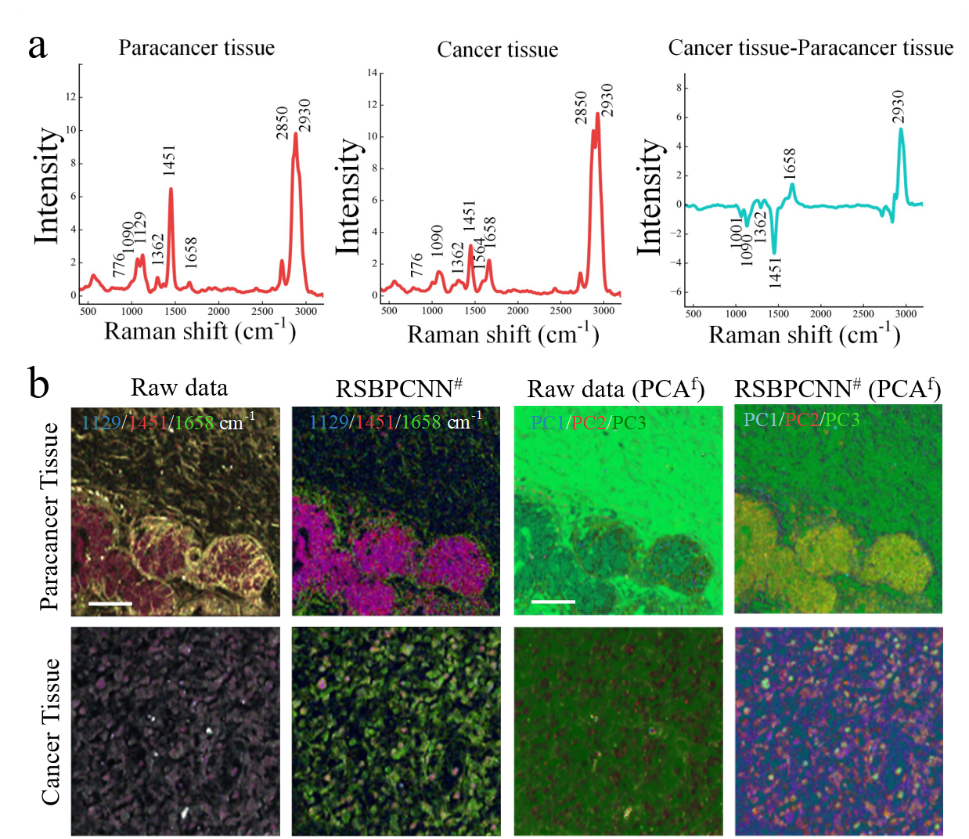
Figure 2. Effects of Raman imaging on pathological tissues
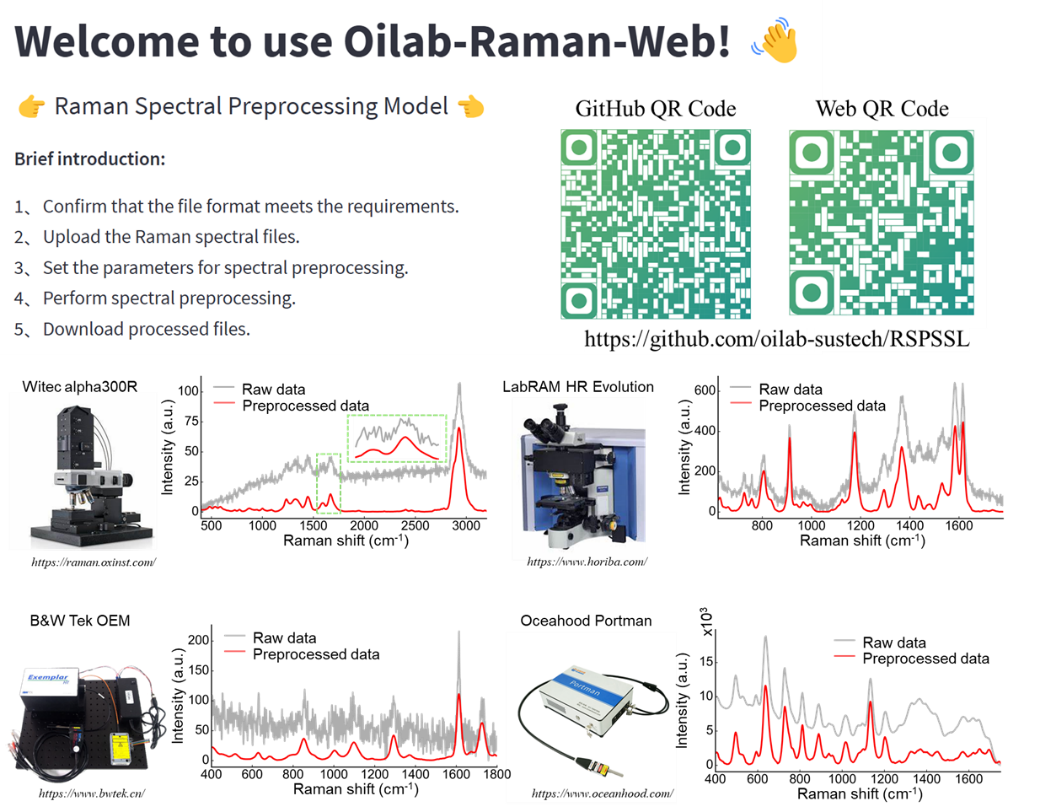
Figure 3. Generalized Raman spectroscopy preprocessing platform
Jiaqi Hu, a Ph.D. student at SUSTech, is the paper’s first author. Research Associate Professor Gina Jinna Chen (co-first author) from the Department of EEE and Chair Professor Perry Ping Shum are the corresponding authors. SUSTech is listed as the primary affiliation and corresponding institution.
This research was supported by the National Natural Science Foundation of China (NSFC), Guangdong Basic and Applied Basic Research Foundation, and the Shenzhen Science and Technology Program.
Paper link: https://doi.org/10.1038/s41377-024-01394-5
Related link: https://github.com/oilab-sustech/RSPSSL
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo ByDepartment of Electronic and Electrical Engineering