In eukaryotic cells, translation initiation and termination are highly regulated biological processes directly impacting protein expression and function. However, existing gene annotation methods and experimental techniques face significant challenges in identifying non-canonical translation sites and predicting novel coding regions. This is particularly true for long non-coding RNAs (lncRNAs), where traditional annotation approaches often fail to capture their latent coding potential.
A research team led by Chair Professor Zefeng Wang from the Department of Systems Biology, School of Life Sciences at the Southern University of Science and Technology (SUSTech) has made a major breakthrough in this area. They study introduces TranslationAI, a deep neural network model that accurately predicts RNA translation initiation (TIS) and termination sites (TTS). Their work reveals a novel regulatory role of codon usage bias in translation termination, providing powerful new tools and insights for understanding post-transcriptional gene regulation.
Their paper, titled “Analysis of RNA translation with a deep learning architecture provides new insight into translation control”, has been published in the journal Nucleic Acids Research.
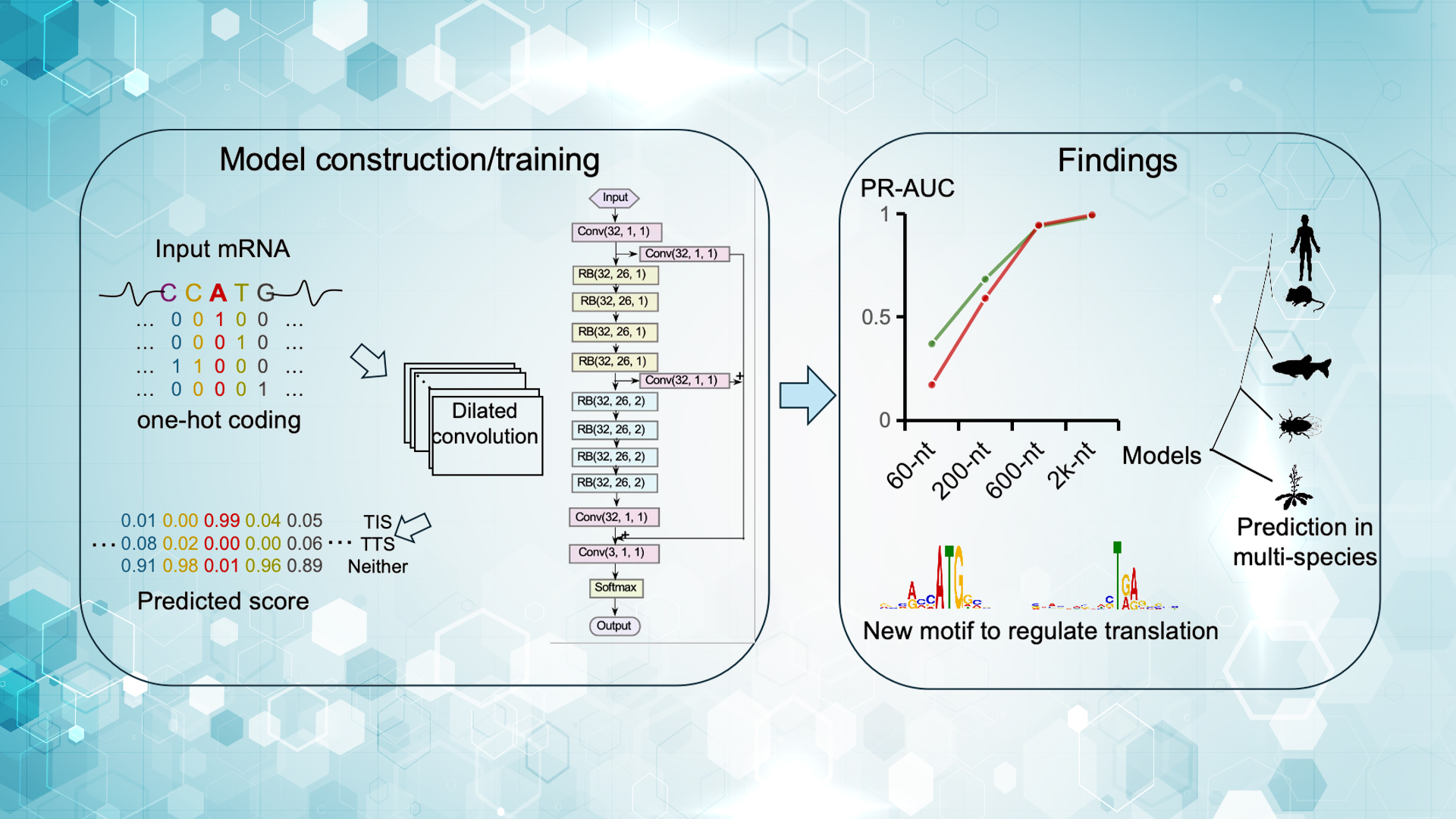
Figure 1. Schematic overview of TranslationAI: Full-length RNA sequences are input into the model, which outputs the probabilities of each site being a TIS or TTS
The team developed TranslationAI, a multilayer dilated convolutional neural network that takes full-length mRNA sequences as input. Without relying on prior features, the model learned hidden sequence patterns governing translation control.
Trained on over 34,000 human reference transcripts, TranslationAI achieved a PR-AUC exceeding 0.99 on independent test datasets, significantly outperforming existing models. Moreover, it demonstrated strong generalizability across a range of model organisms, including yeast, Arabidopsis, and zebrafish, as well as viral genomes such as SARS-CoV-2 and Ebola virus, indicating a high conservation of translation regulatory mechanisms across species.
Beyond accurate prediction of translation sites, the network uncovered a novel regulatory mechanism: codon usage immediately upstream of the stop codon significantly influences translation termination efficiency. Specifically, the presence of C/G-rich synonymous codons upstream of the stop codon enhanced termination efficiency, whereas A/U-rich codons promoted translation readthrough. This finding was experimentally validated across multiple reporters, highlighting the crucial role of “hidden coding information” embedded within mRNA sequences in regulating translation termination.
The team also systematically applied TranslationAI to the human transcriptome, identifying thousands of previously unannotated open reading frames (ORFs), including 673 upstream ORFs (uORFs), 127 downstream ORFs (dORFs), and 3794 novel TIS-TTS pairs in lncRNAs. Several newly predicted ORFs were supported by proteomics data and functional studies, suggesting that non-coding RNAs may harbor extensive hidden coding potential.
TranslationAI has been made publicly accessible as a web tool, enabling researchers to upload RNA sequences and predict their translation initiation and termination sites (related link below).
Drs. Xiaojuan Fan and Tiangen Chang, together with Ph.D. student Chuyun Chen, are the co-first authors of the paper. Chair Professor Zefeng Wang, along with Dr. Xiaojuan Fan, are the co-corresponding authors. Dr. Markus Hafner also contributed to this study.
Paper link: https://doi.org/10.1093/nar/gkaf277
Webpage link to TranslationAI: https://www.biosino.org/TranslationAI/
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yuwen ZENG
Photo ByYan QIU