The development of multicellular organisms involves intricate processes rigorously regulated by numerous genes and signaling pathways across spatial and temporal dimensions. As a classical model organism, Drosophila exhibits remarkable conservation of developmental features with mammals, providing critical insights into embryogenesis and organ formation. While single-cell multi-omics technologies have revolutionized the study of biological development and cellular heterogeneity, spatiotemporal dynamic analysis in Drosophila remained an unresolved gap.
A research team led by Associate Professor Yuhui Hu from the Department of Pharmacology, School of Medicine at the Southern University of Science and Technology (SUSTech), in collaboration with BGI Institute of Life Sciences (BGI), has constructed a 3D spatiotemporal single-cell multi-omics developmental atlas of Drosophila development.
Their study, entitled “A Drosophila single-cell 3D spatiotemporal multi-omics atlas unveils panoramic key regulators of cell type differentiation,” has been published in Cell.
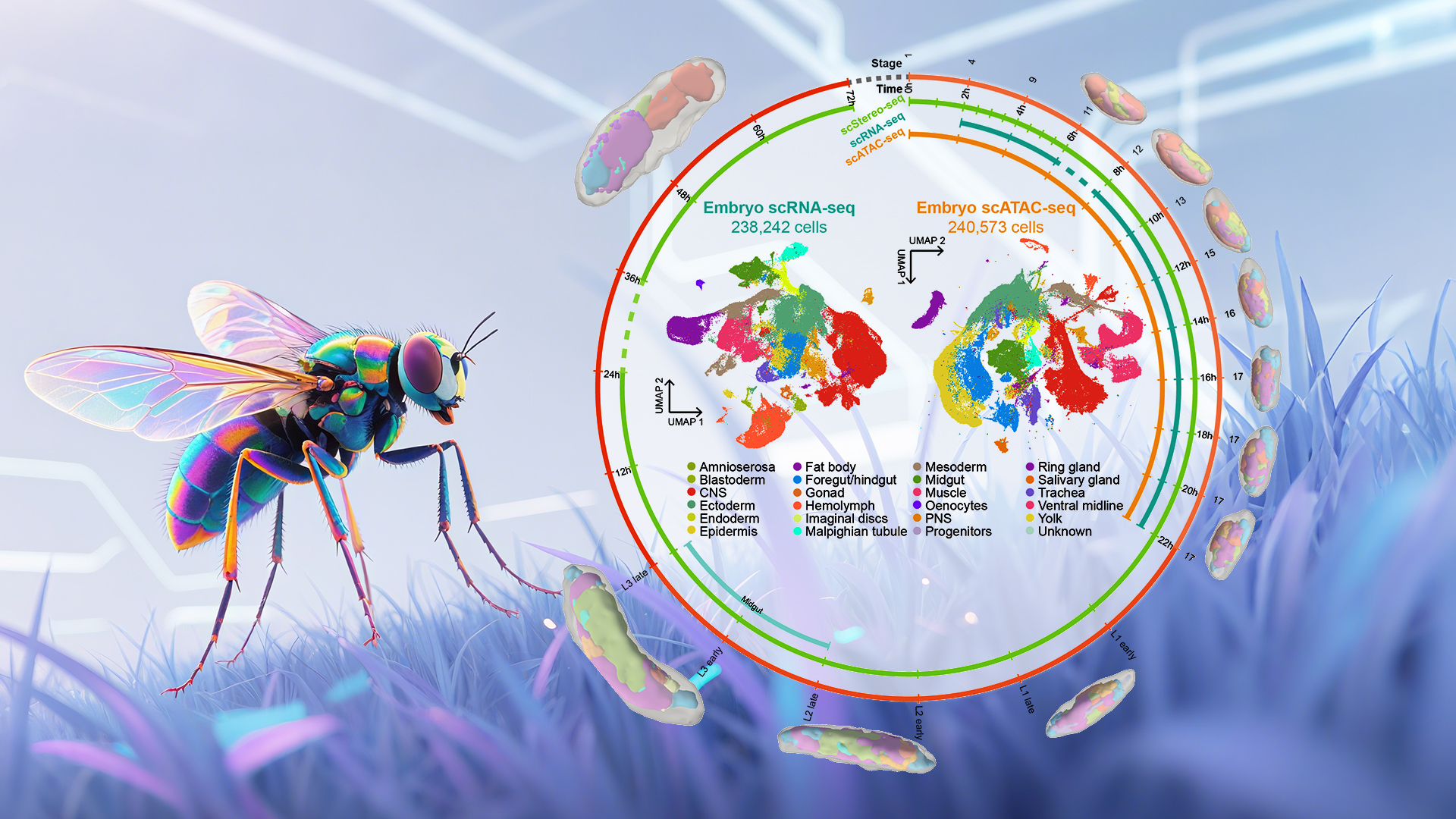
Using multi-omics sequencing technologies, including high-resolution spatial transcriptomics (Stereo-seq), the researchers established the Drosophila developmental atlas Flysta3D-v2 (Figure 1). They integrated multidimensional data, including single-cell spatial transcriptomes, scRNA-seq, and scATAC-seq. Through multimodal data integration, the team constructed continuous whole-organism 3D models revealing detailed trajectories of cell type differentiation (Figure 2). Focusing on the midgut (homologous to the mammalian small intestine), they validated exex as a key regulator of copper cell development via transcription factor screening and mutant analysis.
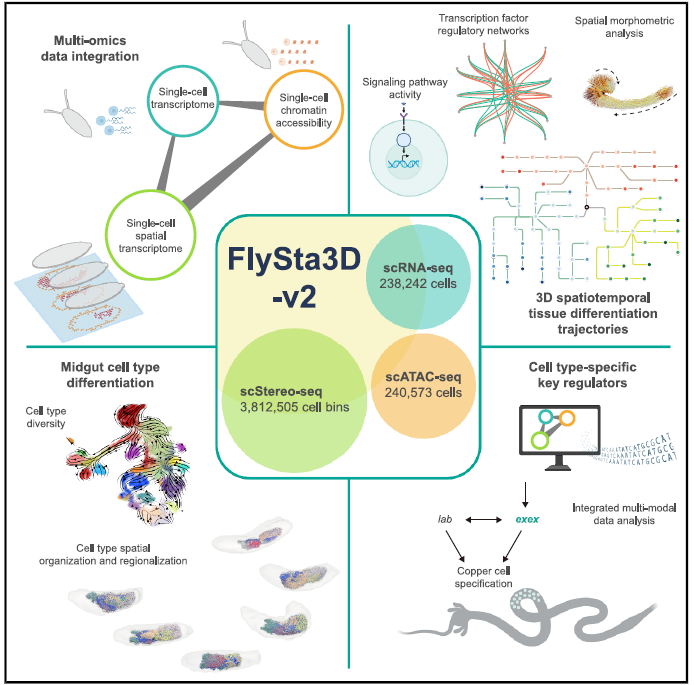
Figure 1. Single-cell 3D spatiotemporal multi-omics atlas of Drosophila development
This study collected samples across embryonic, larval, and pupal stages. Stereo-seq generated 3D single-cell spatial transcriptomes from 43 embryos, 9 larvae, and 5 pupae—totaling >3.8 million single-cell spatial transcriptomes—plus ~240,000 embryonic scRNA-seq and scATAC-seq profiles. Spatial mapping reconstructed 3D transcriptomic atlases of Drosophila tissues, providing a high-resolution molecular map for studying cellular differentiation. This enabled the establishment of regulatory networks of signaling pathways and transcription factors, uncovering spatial patterns of tissue morphogenesis.

Figure 2. Dataset establishment and whole-organism 3D digital reconstruction
Using the embryonic midgut model, the researchers identified functionally specialized cell subtypes, mapped differentiation trajectories/spatial distributions, and analyzed larval/pupal dynamics. Sub-clustering revealed expanded midgut cellular diversity in larvae versus embryos. Regulatory similarities between larval and adult stages indicated early establishment of functional cellular programs (Figure 3).
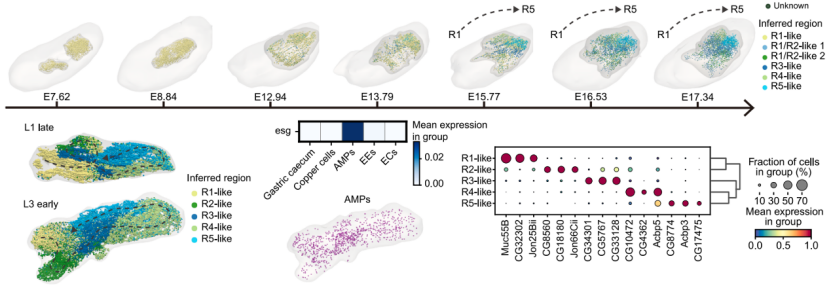
Figure 3. Midgut cell differentiation and functional regionalization
Multimodal analysis identified key transcription factors in midgut development. Exex was determined to regulate copper cell fate determination and metal ion homeostasis. They identified high exex expression and motif activity during early copper cell lineage and experimentally validated its significance in copper cell differentiation.
The Flysta3D-v2 atlas provides an unparalleled resource for Drosophila developmental research. Integrated spatial, transcriptomic, and chromatin accessibility data enable unprecedented resolution in analyzing cell differentiation, tissue patterning, and gene regulatory networks, establishing a foundation for multimodal studies.
This work advances our understanding of tissue development in Drosophila and offers paradigms for studying cellular differentiation mechanisms in other organs. It provides critical insights into developmental molecular mechanisms and disease pathophysiology, suggesting potential therapeutic targets.
Dr. Mingyue Wang from BGI, Research Assistant Professor Qinan Hu from SUSTech, Dr. Zhencheng Tu from BGI, and postdoctoral fellow Lingshi Kong from SUSTech are the co-first authors of the paper. The corresponding authors are Associate Professor Yuhui Hu and Research Assistant Professor Qinan Hu, along with Dr. Xun Xu and Dr. Longqi Liu from BGI. SUSTech is the corresponding affiliated institution.
Paper link: https://doi.org/10.1016/j.cell.2025.05.047
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yifei REN
Photo ByDepartment of Pharmacology