Single-molecule localization microscopy (SMLM) is one of the three major super-resolution imaging techniques. By combining ultra-high spatial resolution with molecular specificity, it offers unique advantages over other fluorescence imaging methods. In recent years, the integration of deep learning into SMLM workflows has greatly improved localization accuracy under low signal-to-noise ratio and high emitter density conditions.
As high-throughput super-resolution imaging for high-content screening becomes a key development direction, existing deep learning-based SMLM methods face a major challenge: their high model complexity often results in long processing delays and heavy computational resource demands, limiting deployment in real-world, large-scale imaging scenarios. While model compression techniques can reduce complexity, they often come at the cost of localization accuracy—an unacceptable trade-off for SMLM applications.

A research team led by Associate Professor Yiming Li from the Department of Biomedical Engineering at the Southern University of Science and Technology (SUSTech) has made significant advances in high-efficiency single-molecule localization analysis.
Their research has been published in the top-tier scientific journal Nature Communications under the title “Scalable and lightweight deep learning for efficient high accuracy single-molecule localization microscopy.”
The research team developed LiteLoc, which combines a lightweight dual-stage feature extractor (coarse and fine) with a scalable, competitive-parallel data processing strategy. On an 8× NVIDIA RTX 4090 graphics processing unit (GPU) cluster, LiteLoc achieved a throughput exceeding 560 MB/s, enabling real-time analysis of high-throughput super-resolution imaging powered by modern sCMOS cameras. This work establishes a new benchmark for balancing localization accuracy and computational efficiency in deep learning-based SMLM, offering an efficient and scalable solution for high-content imaging workflows in the life sciences.
The object feature of conventional images is usually spread across a large region within the image, which requires the training of the whole image for tasks such as image classification or restoration. In contrast, SMLM data can be naturally decomposed into numerous small spatiotemporal blocks. Each block contains all the necessary information about the local molecular events, independent of other areas of the image (Figure 1a). However, most existing deep learning SMLM analysis tools adopt a serial processing mode that underutilizes central processing unit (CPU) and GPU resources.
LiteLoc’s parallel framework maximizes hardware utilization by modularizing the workflow into data loading/preprocessing, network inference, and postprocessing/writing steps, and enables multiple GPUs to compete for and process data simultaneously. As a result, the total analysis time approaches the inference time of a single subprocess.
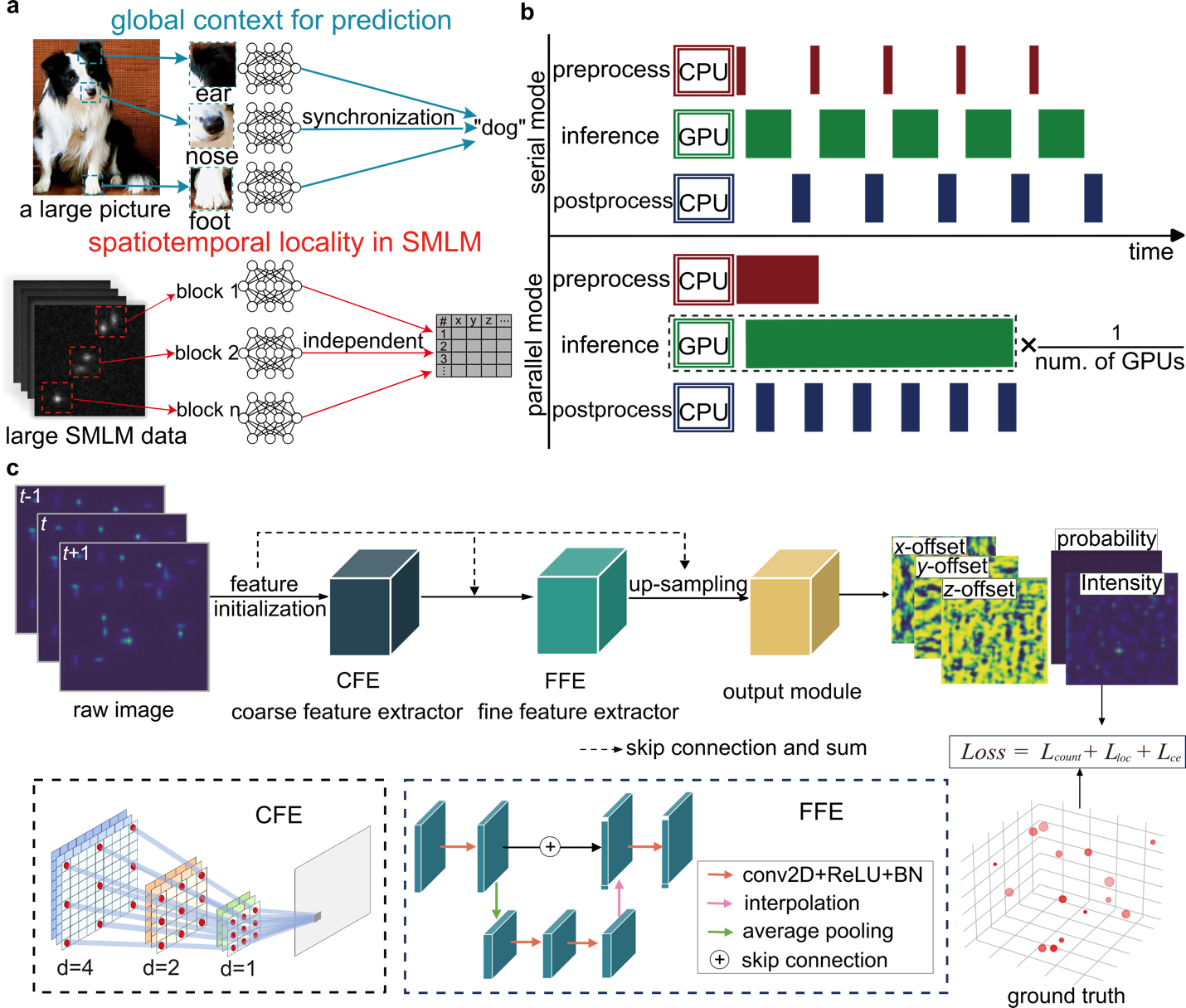
Figure 1. Scalable framework and lightweight architecture of LiteLoc
The LiteLoc network architecture consists of two main components. The first is a coarse feature extractor built from dilated convolution blocks with progressively increasing dilation rates, allowing for a larger receptive field without increasing parameter count or computation. The second is a fine feature extractor designed as a streamlined U-Net for multi-scale information fusion and improved feature reuse (Figure 1c). This design reduces model complexity by an order of magnitude (Figure 2a) while maintaining or exceeding the localization accuracy of the state-of-the-art (SOTA) algorithms such as DECODE (Figure 2c-f). When combined with the parallel processing framework, LiteLoc achieves 567.6 MB/s total analysis speed—surpassing the typical read speed limit of conventional SATA SSDs (500 MB/s).
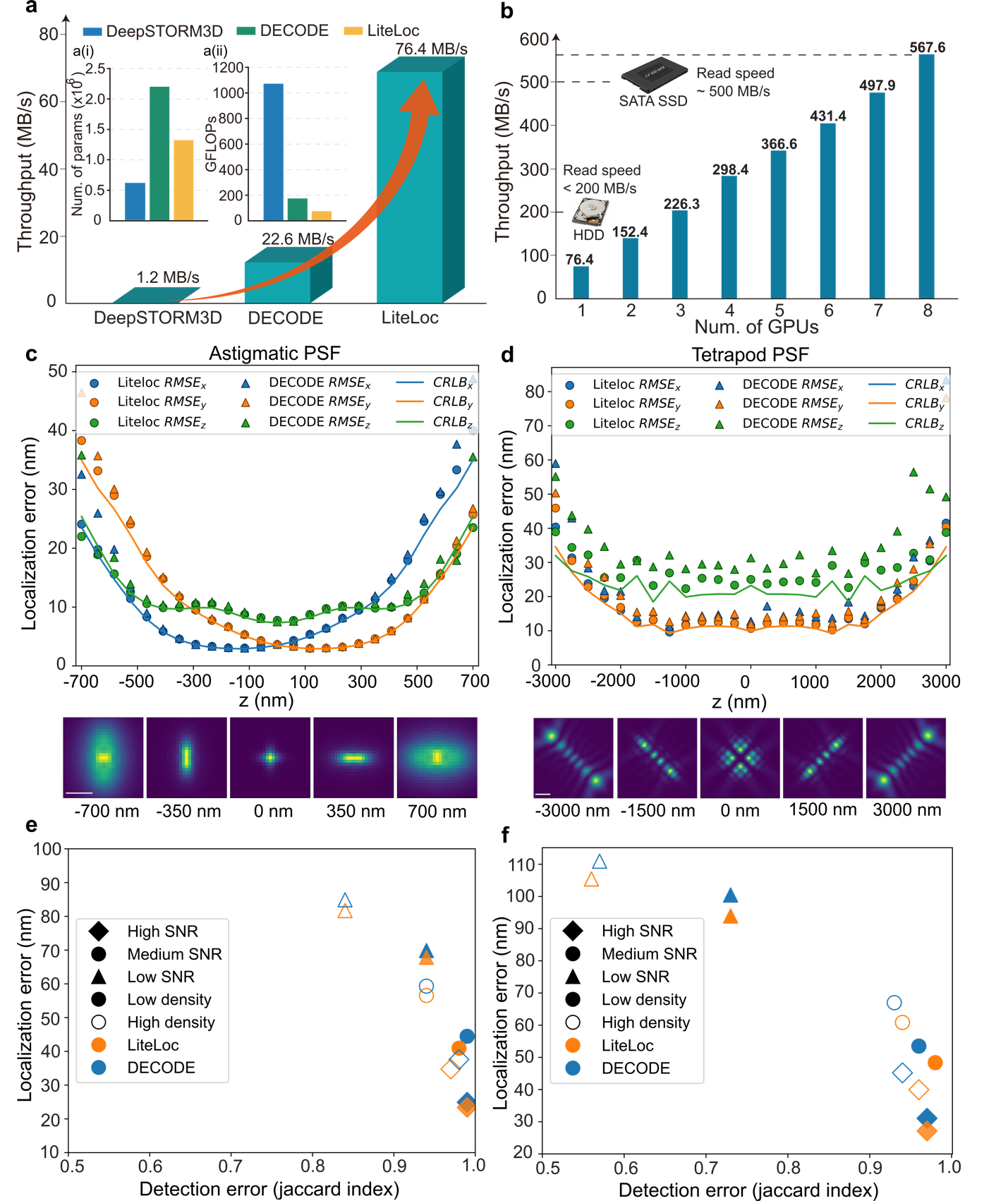
Figure 2. Performance evaluation of LiteLoc on SMLM data with different PSFs
Performance validation was conducted using both astigmatic and 6 μm DMO-Tetrapod point spread functions (PSFs) to image the reference standard nuclear pore complex (NPC) Nup96 in U2OS cells. The known double-ring structure of Nup96 (~107 nm diameter, ~50 nm ring separation) was accurately reconstructed. LiteLoc, DECODE, and DeepSTORM3D all successfully resolved the double ring structure in 3D near the coverslip (Figure 3a-c). However, when imaging with a 6 µm DMO-Tetrapod PSF, the localizations of DeepSTORM3D appeared more dispersed (Figure 3e), and the number of emitters detected were less, as shown in the side view of the cell (Figure 3i).
The super-resolution image reconstructed using DECODE tended to produce grid-like artifacts, while no obvious artifacts were observed in the images reconstructed by LiteLoc (Figure 3e), which demonstrates more precise localization. In terms of analysis speed, under the same hardware conditions, LiteLoc required only 28.8% of the analysis time of DECODE and 1.5% of that of DeepSTORM3D, significantly improving the processing efficiency of single-molecule data.
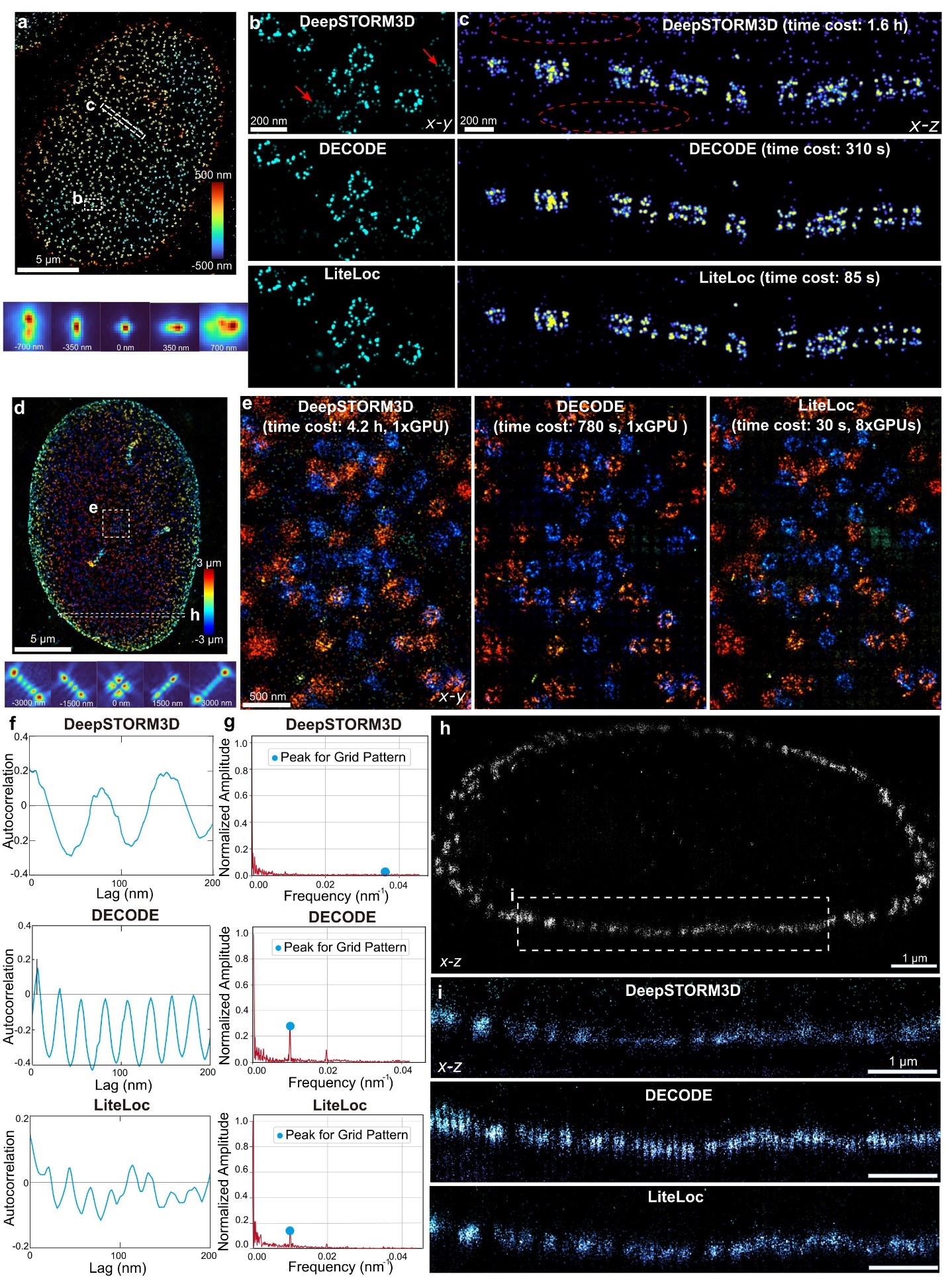
Figure 3. Performance of LiteLoc on biological samples imaged with astigmatic and 6 µ m DMO-Tetrapod PSFs
Conventional deep learning-based SMLM software supports only limited PSF modeling methods, whereas researchers often need to choose different modeling approaches depending on specific imaging requirements. The spline-interpolated PSF is relatively simple and suitable for modeling spatially invariant PSFs; in contrast, the vectorial PSF incorporates multiple optical parameters such as wavelength, refractive index, and numerical aperture, making it more appropriate for scenarios involving depth- and field-dependent aberrations.
LiteLoc supports both PSF modeling approaches for training Reconstruction results using LiteLoc trained with in situ PSF estimation combined with the vectorial PSF model were presented alongside results trained with the spline-interpolated PSF for comparison (Figure 4). LiteLoc trained with the in situ vectorial PSF more accurately matches the actual NPC structure, indicating its superior suitability for compensating depth-dependent aberrations.
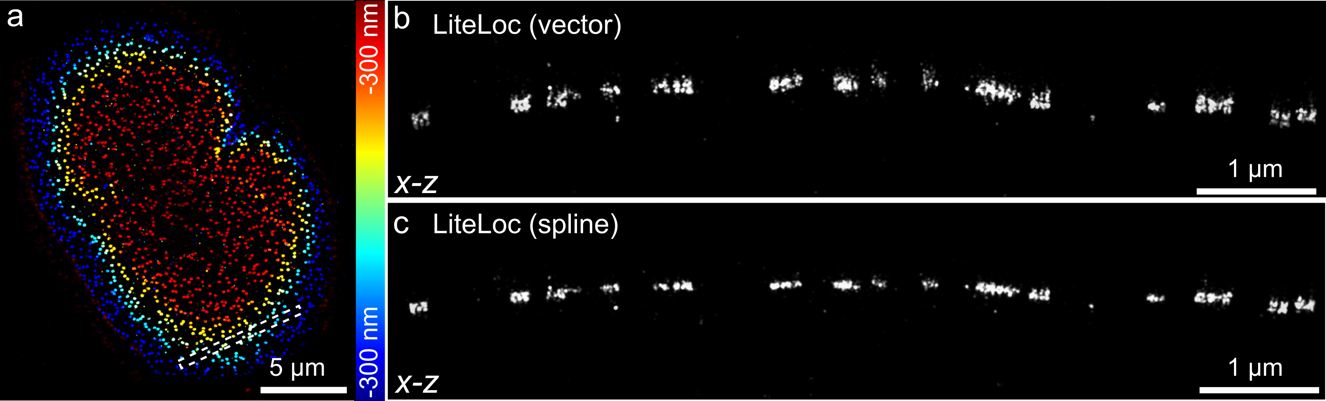
Figure 4. LiteLoc incorporates both spline and vector PSF models for training data generation
This study has introduced a lightweight, scalable deep learning framework tailored for high-throughput SMLM analysis. LiteLoc accelerates inference speed by more than threefold without compromising accuracy, addresses challenges related to PSF complexity, structured noise artifacts, and imaging diversity, and is applicable to a wide range of biological imaging tasks. With its real-time processing capability and low computational demands, LiteLoc can be easily integrated into standard SMLM workflows and combined with downstream modules such as clustering, tracking, and structural reconstruction for fully streamlined super-resolution imaging.
Master’s graduate Yue Fei, postdoctoral fellow Shuang Fu, and Wei Shi are co–first authors of this paper. Associate Professor Yiming Li is the corresponding author.
Paper link: https://www.nature.com/articles/s41467-025-62662-5
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByYiming LI, Adrian Cremin, & Yuwen ZENG
Photo ByDepartment of Biomedical Engineering