S-nitrosylation is a post-translational modification of proteins based on redox reactions, where a nitroso (-NO) group is added to cysteine residues, thereby altering protein conformation, stability, subcellular localization, and biochemical activity to regulate various physiological processes. Quantitatively detecting S-nitrosylation targets in proteins is challenging due to its low abundance and highly dynamic nature in both animals and plants.
Professor Pengcheng Wang’s group from the Institute of Advanced Biotechnology at the Southern University of Science and Technology (SUSTech) recently reported a novel method for studying protein S-nitrosylation, which enables high-precision quantitative detection of S-nitrosylation modifications in plant proteomes. The study also revealed the central role of S-nitrosylation in regulating endoplasmic reticulum (ER) stress responses.
Their work, entitled “FAT-switch-based quantitative S-nitrosoproteomics reveals a key role of GSNOR1 in regulating ER functions”, has been published in the journal Nature Communications.
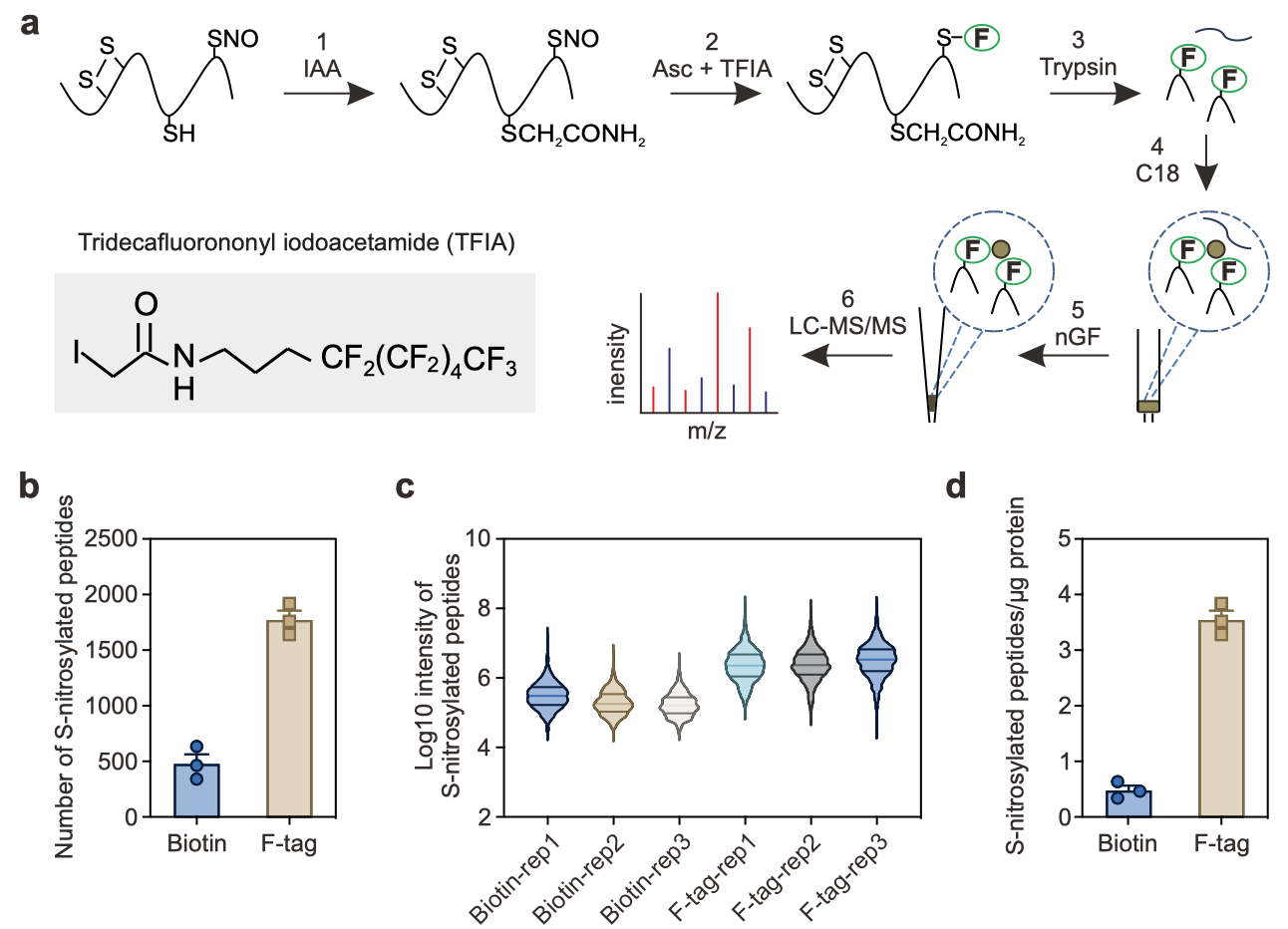
Figure 1. Development of FAT-switch approach for the detection of S-nitrosoproteome in Arabidopsis
Prof. Wang’s team developed a chemical proteomics approach called the Fluorous affinity tag-switch (FAT-switch) for the enrichment and detection of S-nitrosylation peptides. Compared to the traditional Biotin-switch method, the FAT-switch method exhibited approximately 7.4-fold and 10-fold improvement in sensitivity and peptide identification efficiency, respectively. Using this approach, the team quantitatively compared the global S-nitrosylation profiles between wild-type Arabidopsis and the gsnor1/hot5/par2 mutant, which exhibits increased endogenous S-nitrosothiol (SNO) levels.
They identified 2,121 S-nitrosylation peptides in 1,595 protein groups, including several previously unreported S-nitrosylated proteins. Notably, they found that 408 S-nitrosylated sites in 360 protein groups showed significant accumulation in the gsnor1/hot5/par2 mutant. The FAT-switch method represents the most efficient S-nitrosoproteomics method reported to date and enables high-precision quantitative detection of S-nitrosylation modifications in plants.
Based on the proteomic findings, the researchers also discovered several S-nitrosylation targets among ER-related proteins. They revealed that S-nitrosylation of Cys337 in ER OXIDOREDUCTASE 1 (ERO1) causes disulfide rearrangement, leading to enhanced ERO1 activity. This study provides a novel method for studying protein S-nitrosylation and offers valuable resources for investigating S-nitrosylation-regulated ER functions in plants.
In recent years, Prof. Wang’s research group has focused on the development of plant proteomics technologies. They have established in-depth plant phosphoproteomics (Wang et al., Mol Cell, 2018; Lin et al., Nat Commun, 2020), tyrosine phosphorylation (Du et al., Stress Biol, 2022), high-throughput identification of protein kinase substrates (Wang et al., PNAS, 2020), proximity labeling (Zhu et al., JIPB, 2022), and single-cell (type) proteomics methods (Wang et al., JIPB, 2023). They have focused on plant osmotic stress signaling and have elucidated the central role of the RAF-SnRK2 kinase cascade in early osmotic stress and ABA signaling (Lin et al., Nat Commun, 2020; 2021).
Guochen Qin, a former postdoctoral fellow in Prof. Pengcheng Wang’s group and current Head of the Mass Spectrometry Facility at the Institute of Advanced Agriculture Sciences, Peking University, is the first author of this paper. Prof. Pengcheng Wang is the corresponding author. Ph.D. student Menghuan Qu, Profs. Chun-Peng Song and Wei Wang from Henan University, and Prof. W. Andy Tao from Purdue University also participated in this study.
This study was supported by the National Key Research and Development Program and the National Institutes of Health in the United States.
Paper link: https://www.nature.com/articles/s41467-023-39078-0
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByYingying XIA
Photo By