In the long-term coevolution with phages, bacteria have evolved a series of defense systems to defend against phage invasion. Understanding these defense mechanisms is crucial for comprehending bacterial survival strategies and developing innovative biotechnological applications. The study of prokaryotic immune systems, such as the newly elucidated SPARSA system, offers valuable insights into how bacteria protect themselves from genetic threats, with potential implications for genetic engineering and synthetic biology.
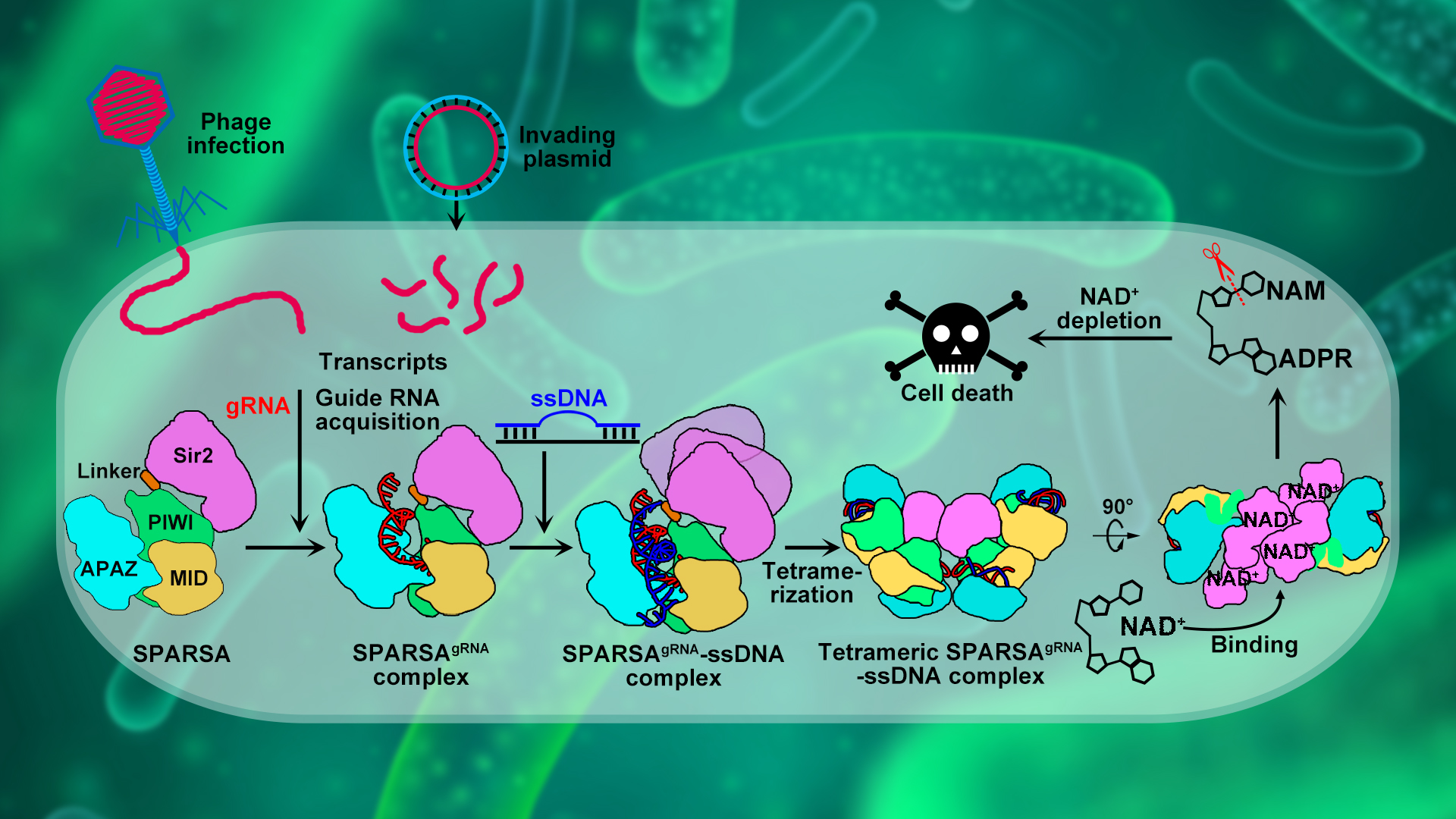
The research team of Associate Professor Ning Jia from the Department of Biochemistry, School of Medicine at the Southern University of Science and Technology (SUSTech) has recently elucidated how bacteria utilize the novel SPARSA system, employing a unique tetramerization-dependent activation mode to combat invasion by exogenous plasmids. The findings deepen our understanding of bacterial defense systems and Ago systems, laying a solid theoretical foundation for the development of biotechnological tools based on the SPARSA system.
Their work, titled “Tetramerization-dependent activation of the Sir2-associated short prokaryotic Argonaute immune system”, has been published in Nature Communications.
The research team has long focused on uncovering the molecular mechanisms by which prokaryotic immune systems, including those in bacteria and archaea, defend against phage and plasmid invasions. In recent years, they have elucidated the mechanisms of various prokaryotic immune systems, such as CRISPR and pAgos, in fending off phage and plasmid attacks (Nature Chemical Biology, 2023; Molecular Cell, 2023, 2019a, 2019b, 2019c; Nature Communications, 2024, 2022; Science, 2020; Cell Research, 2022, 2020; Nature Reviews Molecular Cell Biology, 2021).
Argonaute (Ago) proteins are ubiquitous across all domains of life, from bacteria to humans, and play a key role in RNA interference, a crucial physiological process in eukaryotes. Recent studies have identified a novel class of short prokaryotic Ago (pAgo) proteins in prokaryotes, which are often coupled with NADase domains and are integral in bacterial anti-phage systems. However, the molecular mechanisms underlying their function remain unknown.
The SPARSA (Sir2-domain associated short pAgo) system is a novel type of prokaryotic pAgo system and coupled with a deacetylase (Sirtuin, Sir2) domain, which enables defense by depleting the crucial dinucleotide molecules NAD(P)+ within the cell through its Sir2 domain. To explore the underlying molecular mechanism, the researchers first conducted in vivo experiments that revealed the SPARSA system interferes with the transformation of plasmids, specifically containing ColE1-like origins of replication (ori). They also confirmed that Sir2-APAZ and pAgo form a stable heterodimeric SPARSA complex (Figure 1a, b). Guided by a gRNA strand, this complex recognizes the invading exogenous single-stranded DNA (ssDNA) and subsequently assembles into a tetramer with NAD(P)+ degradation activity (Figure 1b, c). Using cryo-electron microscopy (cryo-EM) single-particle analysis, the team resolved the structures of the inactive SPARSAgRNA-ssDNA monomer and the active SPARSAgRNA-ssDNA tetramer, revealing the molecular mechanisms of SPARSA system assembly, nucleic acid recognition, and NADase activation.
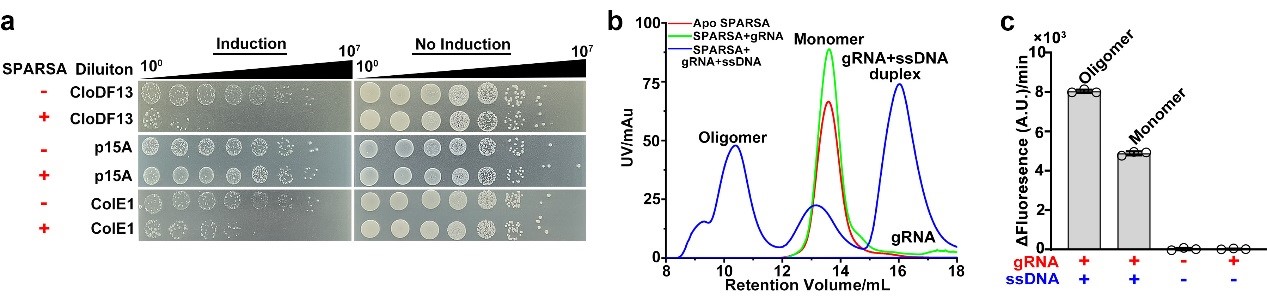
Figure 1. The SPARSA system defends against plasmid invasion and is induced to assemble into an active multimeric complex by ssDNA
The tetrameric SPARSA complex adopts a bowl-shaped architecture with the four Sir2 domains positioned at the center of the structure, while the nucleic acid duplex and other domains are located on the periphery (Figure 2). Compared to the monomeric SPARSA-gRNA-ssDNA complex, the four monomers within the SPARSA-gRNA-ssDNA tetramer can be divided into two distinct conformations. The primary difference between these conformations lies in the relative positions of the Sir2 domains and the flexible regions connecting the Sir2 and APAZ domains. This variation indicates that the Sir2 domains undergo a rearrangement during the tetramerization process of the SPARSA system.
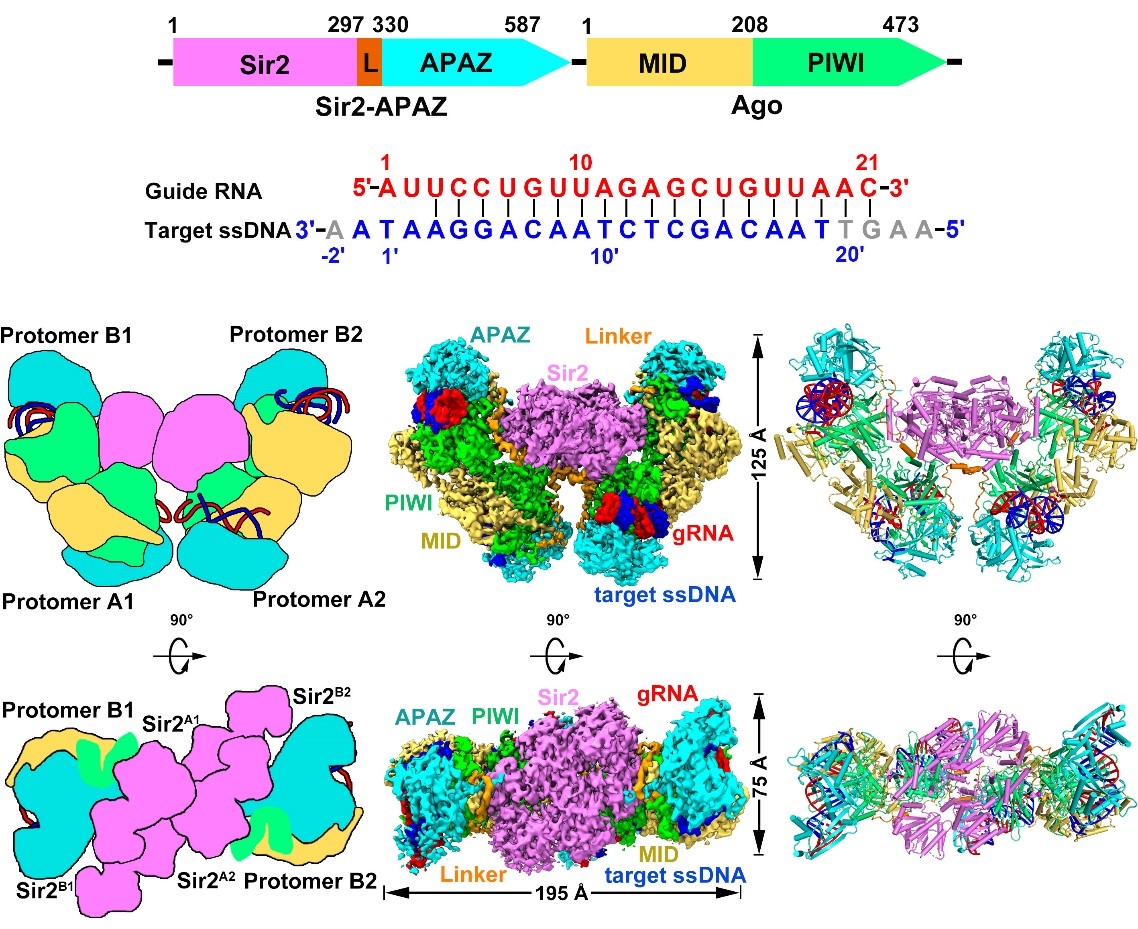
Figure 2. The overall structure of tetrameric SPARSA
Furthermore, the researchers discovered that the SPARSA system contains a specific flexible region within its pAgo protein that can detect whether the system has complementarily bound to ssDNA under the guidance of RNA. By comparing the structure of the SPARSA complex in its nucleic acid-bound and unbound states, they found that this flexible region undergoes significant conformational changes and directly interacts with the bases of the RNA-DNA duplex. Mutating this flexible region completely suppressed the NADase activity of the SPARSA system, indicating that this region plays a crucial role in activating the SPARSA system.
In summary, the team proposed a model (Figure 3) for the processes of SPARSA complex assembly, nucleic acid recognition, and multimeric activation, providing a detailed explanation of the molecular mechanisms by which the SPARSA system defends against plasmid invasion. These findings enhance our understanding of the prokaryotic SPARSA defense system and establish a molecular foundation for developing biotechnological tools based on the SPARSA system.
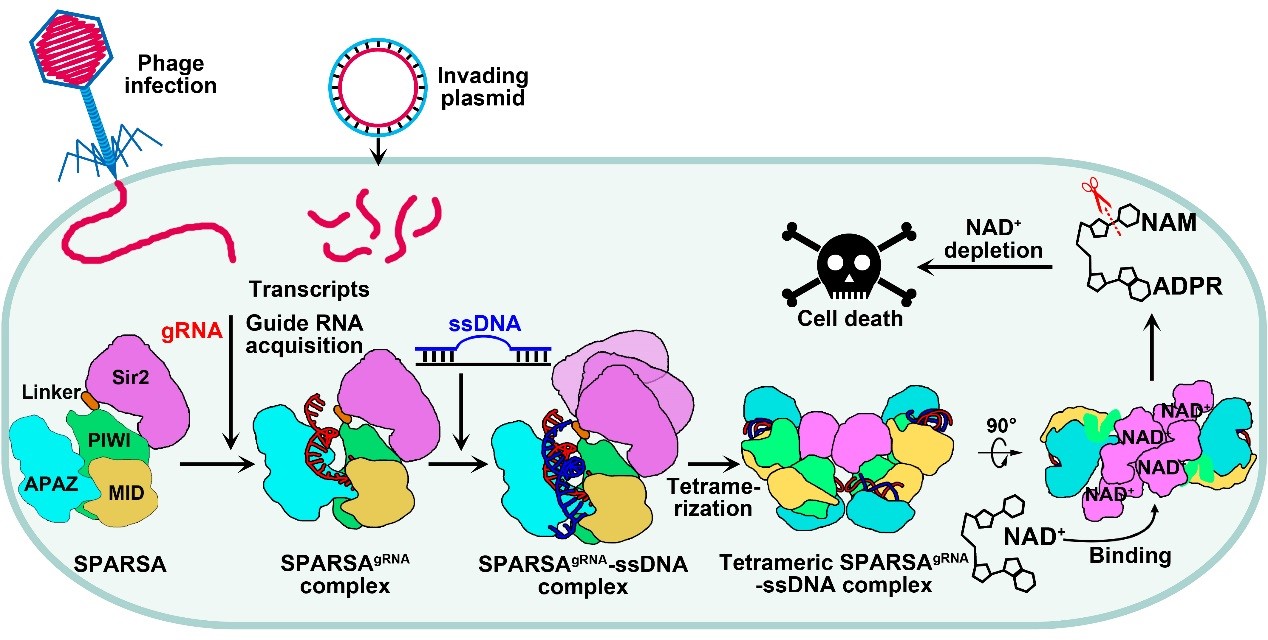
Figure 3. Proposed mechanistic model for oligomerization-dependent activation of SPARSA NADase activity in response to invading mobile genetic elements
Postdoctoral researchers Ning Cui and Jun-Tao Zhang at SUSTech are the co-first authors of this paper. Associate Professor Ning Jia is the corresponding author.
Paper link: https://doi.org/10.1038/s41467-024-52910-5
Proofread ByAdrian Cremin, Yingying XIA
Photo ByDepartment of Biochemistry