With RNA being the core carrier of genetic information in life, understanding how RNA and small interfering RNA (siRNA) transmits responses to external stress may be better understood following new research led by Southern University of Science and Technology.
Chair Professor Hongwei Guo (Biology) led a multinational research effort to improve the understanding of how RNA interference works in conjunction with the stress response. Their research on this matter, as it related to plant RNA, was published in the high impact academic journal Nature (IF = 43.07), under the title, “Plant 22-nucleotide siRNAs mediate translational repression and stress adaptation.”
RNA interference regulates the target RNA gene through small pieces of RNA that tend to be between 21-24 nucleotides (nt) long. Small RNAs, such as miRNAs and siRNAs, are essential for animal and plant growth. Each plant’s small RNA has a different length, with the length determining the biological function.
The paper identified a large number of endogenous 22nt siRNAs in mutants, which revealed an essential biological function in the model plant, Arabidopsis. It measured stress adaptability and translation inhibition, representing a breakthrough in small RNA in plants. It is an advance on their previous work published in Science in 2015.
The research team in their 2020 paper found that when the model plants lack a specific RNA degradation pathway and do not have DCL4, one of the four Dicer-like (DCL) proteins, there are severe growth and development defects. They could also determine that the production of 22nt siRNAs is dependent on RNA-dependent polymerase 6 (RDR6) and DCL2.
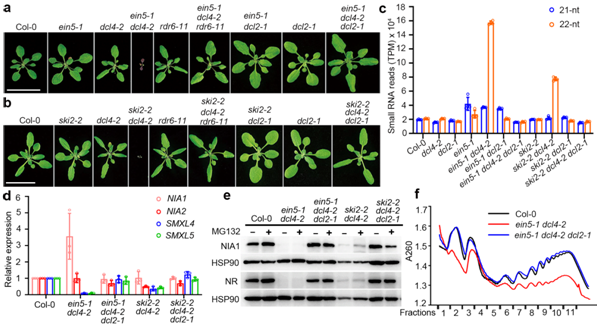
Figure 1. The ein5 dcl4 and ski2 dcl4 double mutants produce large amounts of 22nt siRNA and mediate translational inhibition.
The team also found that the 22nt siRNAs produced by two nitrate reductase genes (NIA1 and NIA2) involved in nitrogen metabolism account for about half the 22nt siRNAs. However, most of the genes that produce 22nt siRNA did not show significantly reduced mRNA expression.
The findings of this research are significantly different from 21nt siRNA from the previous work, so it is believed that 22nt siRNA could have a different mode of action. By performing a combination of biochemical and genetic assays, they found, for the first time, that after 22nt siRNA binds to the AGO1 protein, it leads to the translation inhibition of their cognate mRNAs. Meanwhile, the AGO1-associated 22nt siRNA induces the secondary amplification of small RNA, increasing the amount of 22nt siRNA when DCL4 activity is lacking.
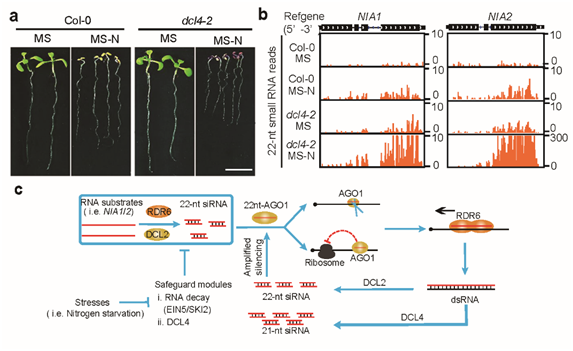
Figure 2: 22nt siRNA is involved in nitrogen stress response and its working model.
The research team examined the biological function of 22nt siRNA under natural conditions. They found that when nitrogen nutrition is limited, plants exhibited growth-inhibiting phenotypes, and accumulated a large amount of 22nt siRNAs, including those from NIA1/2. They also found that plant translation generally decreased under nitrogen deficiency.
The research team hypothesized that plants respond to the stress of extreme nitrogen deficiency by reducing protein use and increasing 22nt siRNA production. This response reduces the level of NIA proteins, which is responsible for reducing nitric acid to nitrous acid. Nitrous acid is further reduced to ammonium ions, which are essential raw materials for the synthesis of amino acids.
The research team concluded that the rarely studied 22nt siRNA provides a new mechanism for dealing with external environmental stresses. Plants can reduce the efficiency of protein translation and reduce energy consumption by temporarily inhibiting their growth state. The activation of their stress response ensures their survival. The translation regulation mechanism of the proteins could involve their response to different environmental stresses and may be vital regulatory nodes.
SUSTech is the first communication unit, with Hongwei Guo is the correspondent author. Visiting student from Peking University (PKU) Huihui Wu and Research Assistant Professor Bosheng Li are the co-first authors. Additional contributions came from the University of Tokyo, Japan Science and Technology Agency (JST), Beijing Forestry University, and Northeast Forestry University.
This work was strongly supported by the National Natural Science Foundation of China, the National Key R & D Program of the Ministry of Science and Technology, the Guangdong Innovation and Entrepreneurship Team Fund, and the Shenzhen Science and Technology Commission Fund.
Paper link: https://www.nature.com/articles/s41586-020-2231-y
Proofread By
Photo By