Mammalian genomes are diploid, comprising of two haploid genomes from each paternal or maternal origin. SUSTech scholars have taken a new approach to the study of the diploid genomic architecture by applying multi-omic data to characterize the 3D diploid genome structures.
SUSTech Associate Professor Wenfei Jin (Biology) led his research group to make significant progress in deciphering the diploid genome architecture using a hybrid mouse system. Their research outcomes were published in the high-impact academic journal, Genome Research (IF = 11.09), under the title of “Diploid genome architecture revealed by multi-omic data of hybrid mice.”
Professor Jin said: “The hybrid genome contains a lot of segregating sites, which greatly facilitates how we infer the paternal haploid and maternal haploid of the hybrid genome. With that, we could construct the 3D genome at the haploid level and analyze its impact on gene regulation.”
Most studies solely investigated the average chromatin architectures without considering the differences between homologous chromosomes. It is in part because genetic variations in general individuals could not provide sufficient segregating sites for investigating allele-specific interactome. Characterizing the 3D diploid genome structures remains challenging, and knowledge about the principles of the 3D nucleus organization is limited.
The research group found that inter-chromosomal interaction patterns between homologous chromosomes are similar. Their similarity is highly correlated with their allelic co-expression levels. Reconstructing the 3D nucleus showed the distance from the homologous chromosomes to the center of the nucleus is almost the same. The inter-chromosomal interactions at centromere-ends are significantly weaker than those at telomere-ends. That finding suggested that they are located in different regions within the chromosome territories.
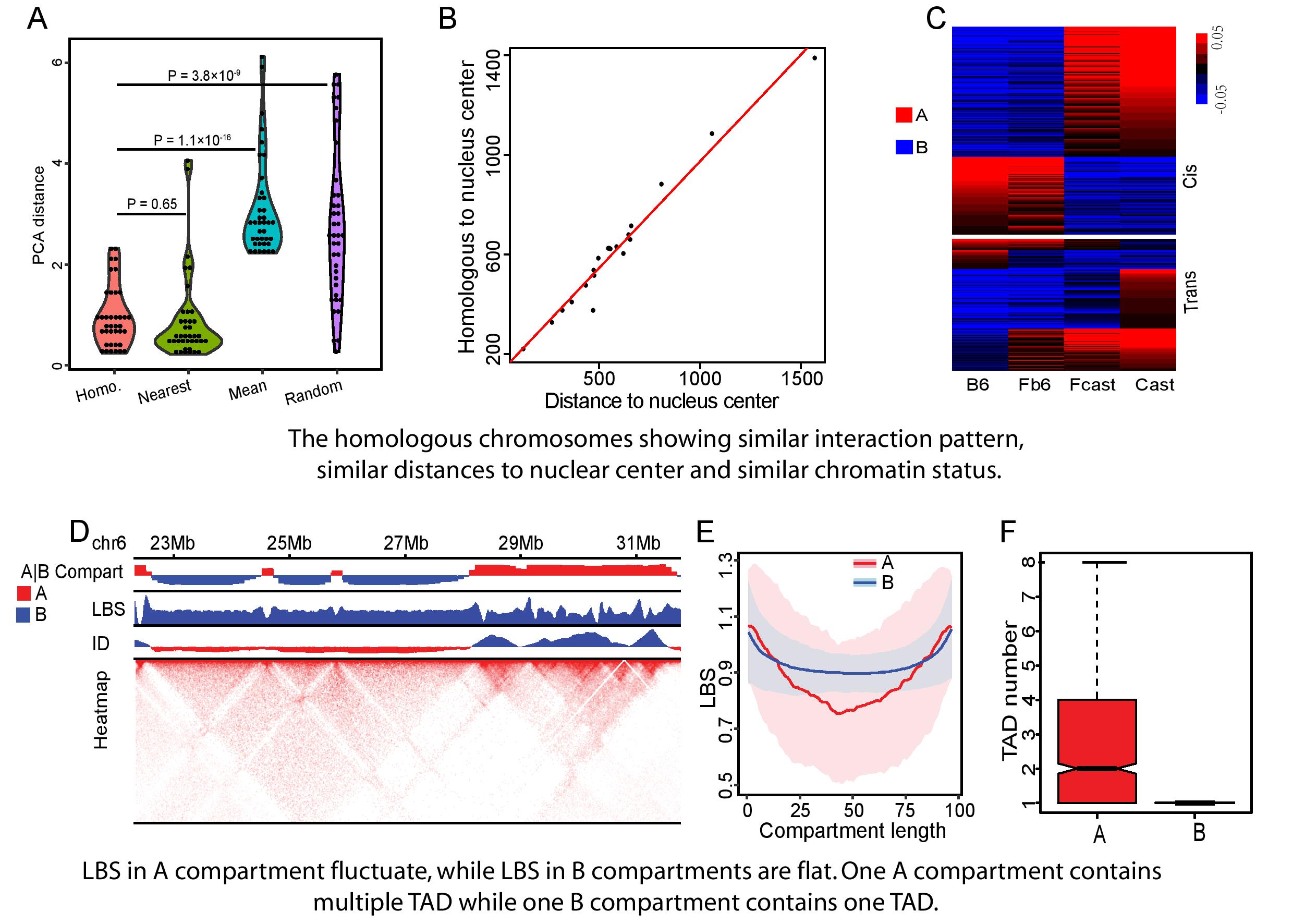
Most of the A|B compartments or topologically associated domains (TADs) are consistent between B6 and Cast. The research group also found that 58% of the haploids in hybrids maintain their parental compartment status at B6/Cast divergent compartments due to cis-effect. About 95% of the trans-effected B6/Cast divergent compartments converge to the same compartment status potentially due to a shared cellular environment.
These results show that differentially expressed genes between two haploids in hybrid mice systems are associated with either genetic or epigenetic effects. The multi-omics data from the hybrid mice have provided haploid-specific information on the 3D nuclear architecture and a rich resource for further understanding the epigenetic regulation of haploid-specific gene expression.
The first author Dr. Zhijun Han and the co-corresponding author Professor Wenfei Jin are from SUSTech. Dr. Keji Zhao from the National Institute of Health (NIH) co-designed this study and is a co-corresponding author. Additional contributing authors include SUSTech Professor Wei Chen and Southeast University Professor Chengqi Lin.
They received financial support from the National Key R&D Program of China, Shenzhen Science and Technology Innovation Committee, and the start-up fund from Southern University of Science and Technology.
Link of the research article: https://pubmed.ncbi.nlm.nih.gov/32759226/
Proofread ByYingying XIA
Photo ByDepartment of Biology, Yan QIU