In eukaryotes, transcription by RNA polymerase II (Pol II) involves initiation, elongation, and termination. The co-transcriptional processes, including mRNA capping, splicing, cleavage, and polyadenylation, turn nascent RNAs into mature mRNAs, which are eventually exported to the cytoplasm. Pre-mRNA splicing occurs mainly co-transcriptionally and is tightly coupled with Pol II elongation. Although the recruitment of splicing factors and the structure and assembly process of the spliceosome have been resolved relatively clearly, the splicing kinetics, co-transcription, and 3’end processing still need to be further studied.
Recent progress in full-length RNA sequencing permitted by single-molecule long-read sequencing platforms (PacBio and Nanopore in particular) have furthered our knowledge on transcriptional regulation by enabling simultaneous detection of elongation and splicing on the same nascent RNA molecule, it allows a comprehensive analysis in identifying full-length splice isoforms, and also splicing kinetics and the coordination of splicing patterns across the multi-intron genes in the nascent RNA.
Based on the full-length sequencing technology, Associate Professor Jixian Zhai’s team from the Institute of Plant and Food Science at the Southern University of Science and Technology (SUSTech) developed Full-Length Elongating and Polyadenylated RNA sequencing (FLEP-seq). This is a full-length nascent RNA sequencing method that simultaneously detects RNA polymerase II position, splicing status, polyadenylation site (PAS), and poly(A) tail length at a genome-wide scale. Their study, entitled “FLEP-seq: simultaneous detection of RNA polymerase II position, splicing status, polyadenylation site, and poly(A) tail length at genome-wide scale by single-molecule nascent RNA sequencing” has been published in the journal Nature Protocols.
The author described a detailed protocol of FLEP-seq for library production and sequencing (Figure 1) and a complete bioinformatics pipeline from raw data processing to downstream analysis. This method can capture both the nascent elongating and polyadenylated transcripts, simultaneously detects the Pol II position, splicing status, polyadenylation sites, and poly(A) tail length on single molecules. With the splicing status, pol II position, and poly(A) information, users can analyze co-transcriptional splicing kinetics, post-transcriptional intron retention, different PAS isoforms, the overall landscape of poly(A) tail length, and correlation between splicing and poly(A) tail length at the gene level (Figure 2).
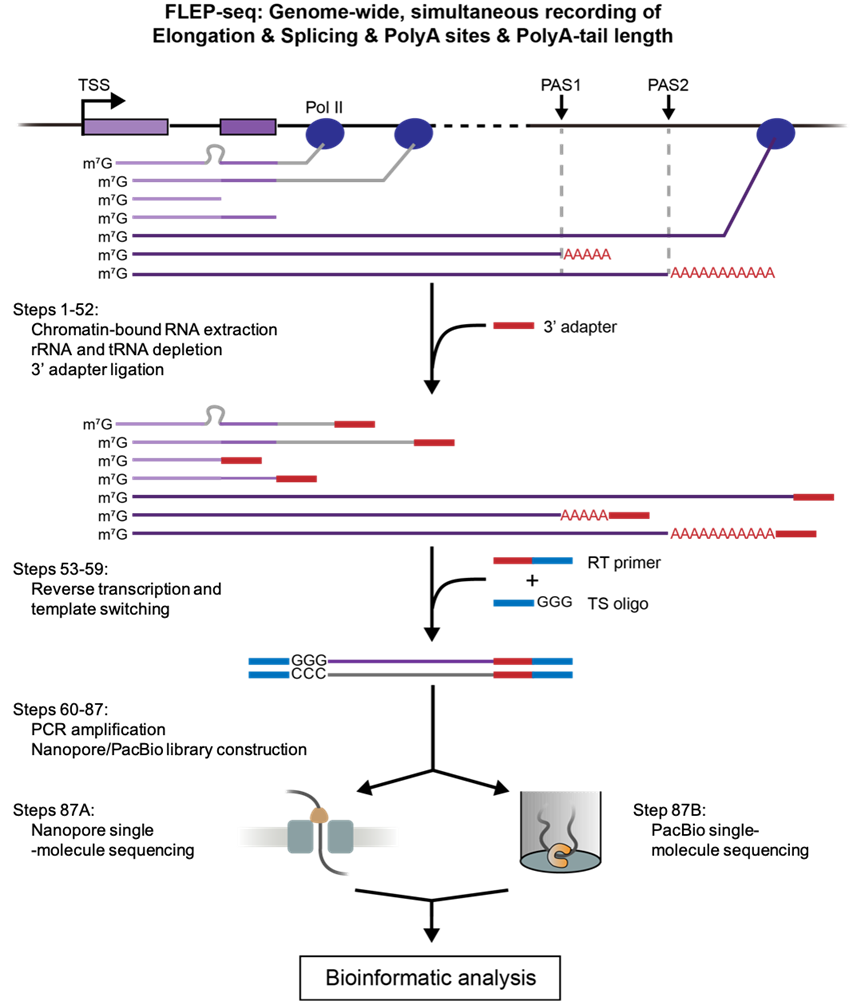
Figure 1. Overview of FLEP-seq protocol
By taking advantage of the comprehensive information on a full-length nascent RNA molecule, FLEP-seq can uniquely reveal the complexity among elongation, splicing, polyadenylation sites, and poly(A) tail length, providing more insights into the RNA processing and transcriptional regulation.
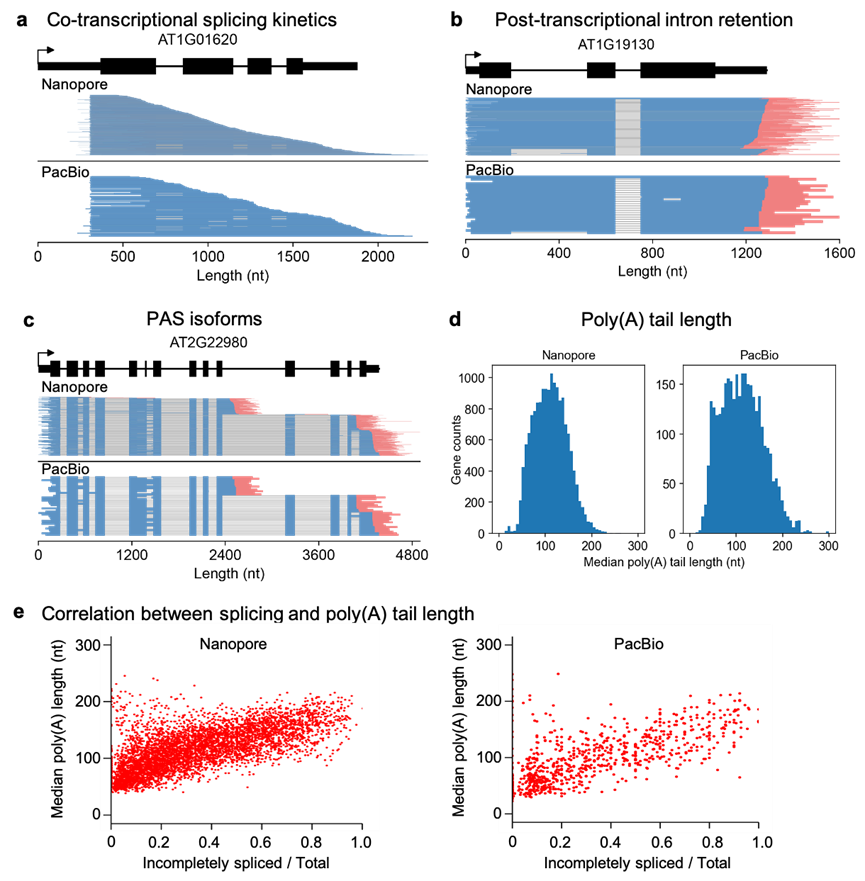
Figure 2. Application of FLEP-seq
Associate Professor Jixian Zhai is the corresponding author of the paper. Research Assistant Professor Dr. Yanping Long and Dr. Jinbu Jia are the co-first authors. This research was supported by the National Key R&D Program of China Grant, the Program for Guangdong Introducing Innovative and Entrepreneurial Teams, the Shenzhen Sci-Tech Fund, and the Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes.
Related links:
Nature Protocols paper: https://www.nature.com/articles/s41596-021-00581-7
Jixian Zhai’s group website: http://www.jixianzhai.org/
Proofread ByAdrian Cremin, Yingying XIA
Photo By