Light signals are essential for plant development and growth. Plants evolve different receptors to precisely sense the different wavelengths of sunlight and thereby initiate photomorphogenesis. The E3 ligase COP1-SPAs complex acts as a central repressor during photomorphogenesis and can promote the degradation of various photomorphogenesis-promoting factors through ubiquitination. Light-activated photoreceptors, such as the UV-B receptor UVR8, can suppress the activity of COP1-SPAs to promote photomorphogenesis. However, the structural mechanism of photoreceptor-mediated suppression of COP1-SPAs is still largely unknown.
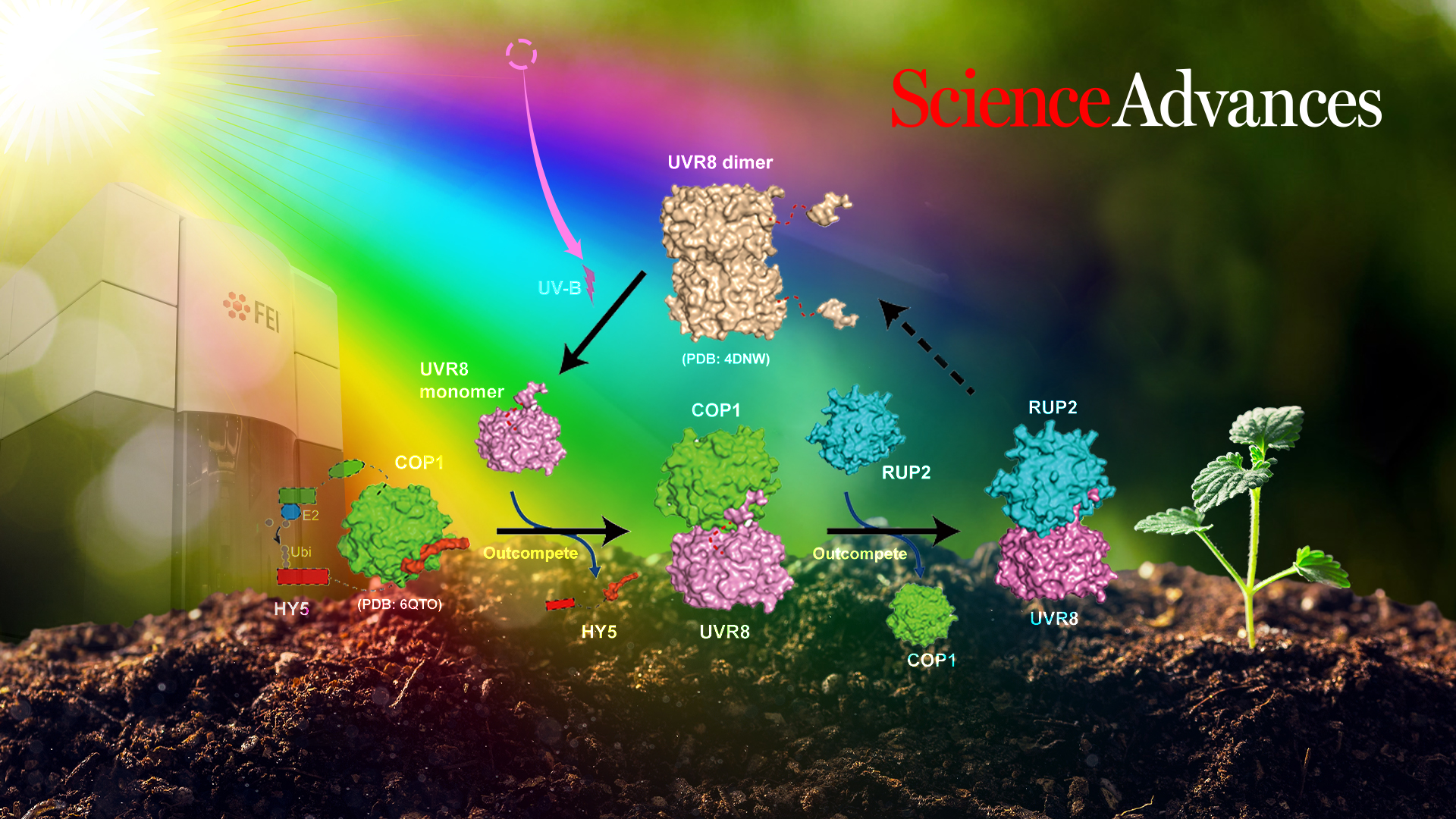
Professor Xing Wang Deng from the Department of Biology at the Southern University of Science and Technology (SUSTech), in collaboration with Professor Ping Yin of Huazhong Agricultural University (HZAU), have recently released an article that shows how the UV-B receptor UVR8 physically interacts with the COP1-SPAs complex and how UVR8 competitively inhibits the interaction of COP1-SPAs and its target HY5.
Their paper, entitled “Structural insight into UV-B-activated UVR8 bound to COP1,” was published in Science Advances, a multidisciplinary journal covering all areas of science.
In this work, the researchers succeeded in purifying a homogeneous and stable COP1-SPA4 complex suitable for subsequent biochemical experiments. UV-B triggered the formation of UVB-COP1-SPA4 and was successfully reconstituted in vitro. Monomeric UVR8 induced by UV-B irradiation could be bound to COP1-SPA4 to form a UVR8-COP1-SPA4 complex. Subsequently, the structure of the UV-B-activated UVR8-COP1 complex was determined by cryo-electron microscopy.
The structure indicated that the interaction of COP1 and UVR8 is mediated by two distinct interfaces. Further biochemical analysis confirmed that these two interfaces are essential for the interaction of UVR8 and COP1 and the dissociation of HY5 from COP1-SPA4. It also revealed the structural features of the core domain of activated UVR8, which were used to identify UVR8W285A, D96N, D107N and UVR8W285A, G01S, two novel activated monomeric UVR8 variants. In addition, the investigators found that RUP2 dissociated UVR8 from the COP1-SPA41-UVR8 complex and facilitated its redimerization and inactivation.
This study elucidates the structural mechanism underlying the assembly and disassembly of UVR8-COP1-SPA4 in UV-B signaling.
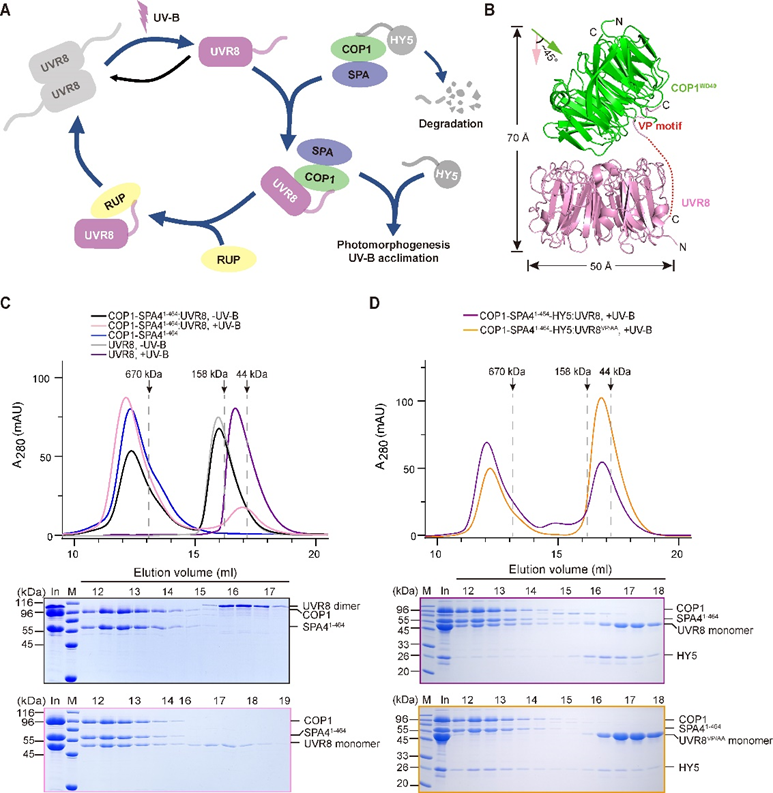
Figure 1. Reconstitution of the COP1-SPA4-UVR8–mediated UV-B signaling pathway
Yidong Wang, Lixia Wang, and Zeyuan Guan from HZAU are the co-first authors of this paper. Prof. Ping Yin from HZAU and Prof. Xing Wang Deng from SUSTech are the co-corresponding authors. Research Assistant Professor Jian Li from the Department of Biology at SUSTech also contributed to this study.
This work was supported by the Ministry of Science and Technology (MOST), National Natural Science Foundation of China (NSFC), Wuhan Applied Foundational Frontier Project, Foundation of Hubei Hongshan Laboratory, and SUSTech Presidential Funding. The Cryo-EM data were collected by the Cryo-Electron Microscopy Center at SUSTech.
Paper link: https://www.science.org/doi/10.1126/sciadv.abn3337
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By