Drosophila melanogaster has long been widely used in biomedical research in genetics and developmental biology. Studies in its various physiological features, including embryogenesis and organogenesis, provided critical insights into conserved processes in mammals. The formation and organization of embryos and tissues are complex procedures involving the orchestration of multiple genes and signal pathways over space and time. To comprehensively understand the development of an embryo and its tissues, it is vital to integrate the spatial and temporal information of gene expression at the genome level.
Associate Professor Yuhui Hu’s team from the School of Medicine at Southern University of Science and Technology (SUSTech), together with collaborators from BGI Shenzhen, recently completed the first 3D reconstruction of spatial transcriptomes of developing Drosophila with high resolution and sensitivity.
This work, entitled “High-resolution 3D spatiotemporal transcriptomic maps of developing Drosophila embryos and larvae,” was published in the journal Developmental Cell. It was also selected as the cover story of the issue.
Drosophila melanogaster has a distinctive but relatively shorter life cycle (Figure 2). Due to the small scale of Drosophila embryos (about 0.5 mm), investigations into spatiotemporal gene expression patterns during its development had been limited to a few genes at a time. Resolving the spatial patterns of gene expression in Drosophila embryos relied largely on in situ hybridization (ISH). Owing to the inherent drawbacks of ISH, such as bias in probe selection and difficulty in multiplexing, a genome-wide study in spatiotemporal patterns of gene expression in Drosophila embryos has been lacking.
Figure 2. Life cycle of Drosophila melanogaster
The newly developed Stereo-seq (SpaTial Enhanced Resolution Omics-sequencing) platform achieved significant breakthroughs in resolution and sensitivity, making it possible to apply chip and sequencing-based spatial transcriptomic techniques to Drosophila embryos. Utilizing Stereo-seq, the researchers performed effective in situ mRNA capture in cryosection slices of Drosophila embryos and larvae.
Based on the sequencing data, they then performed spatial visualization, bin clustering, and tissue annotation to generate 2D spatial transcriptomic maps of embryo and larva sections. By combining all the sections of a sample, the authors were able to rebuild 3D spatial transcriptomes for the first time on Drosophila embryos and larvae (Figure 3).
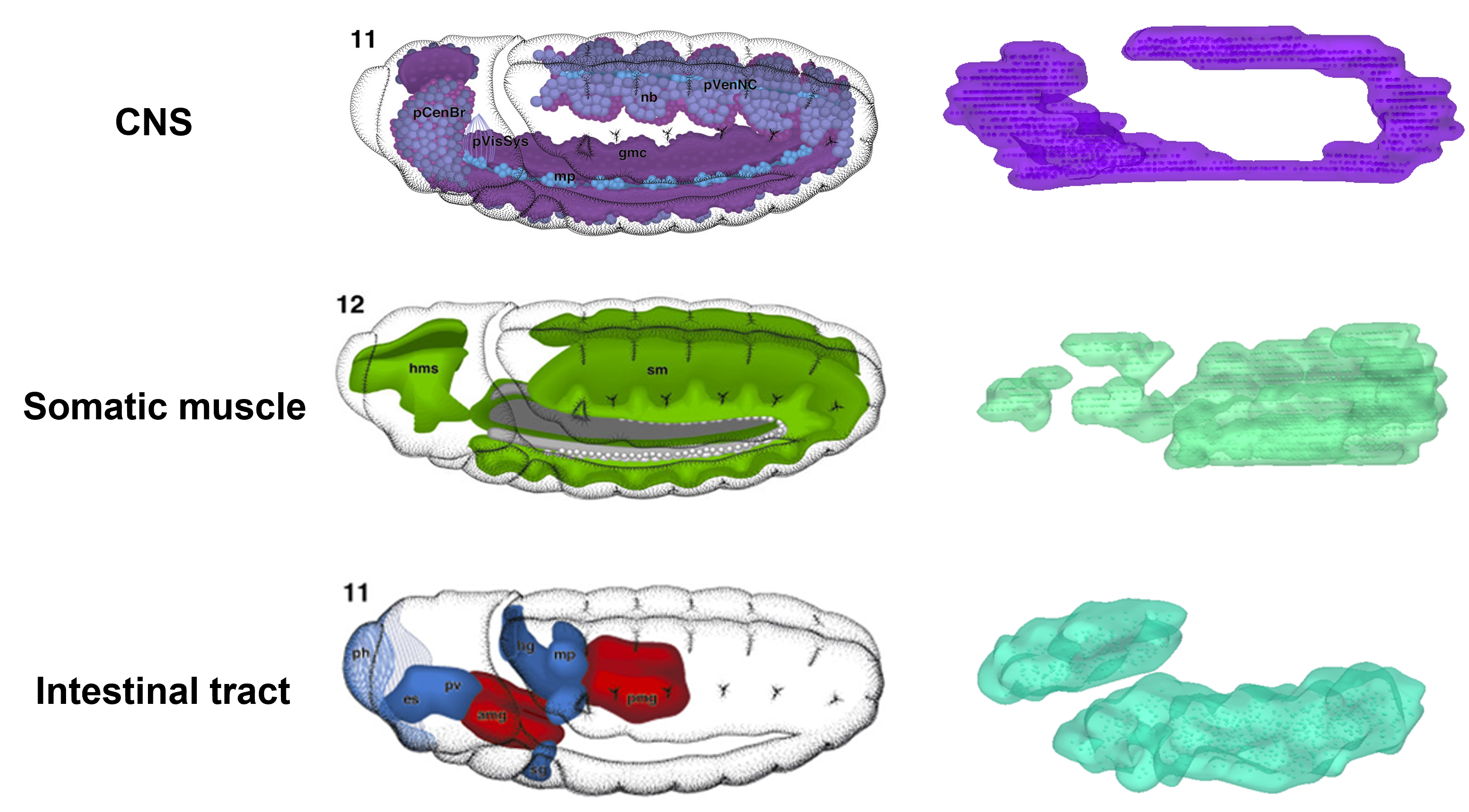
Figure 3. 3D reconstruction of embryonic tissues based on Stereo-seq sequencing data
Analysis of these high-resolution 3D spatial transcriptomes revealed the formation of delicate substructures in tissues like the digestive tract. Further research uncovered spatiotemporal dynamics of cell state changes and transcription factor regulons during development. This study opens exciting opportunities for dynamic analysis on 3D spatiotemporal transcriptomes at genome-scale and whole-organism levels (Figure 4). This provides new angles for systematic studies of gene regulatory networks during the development of embryos, tissues, and diseases.
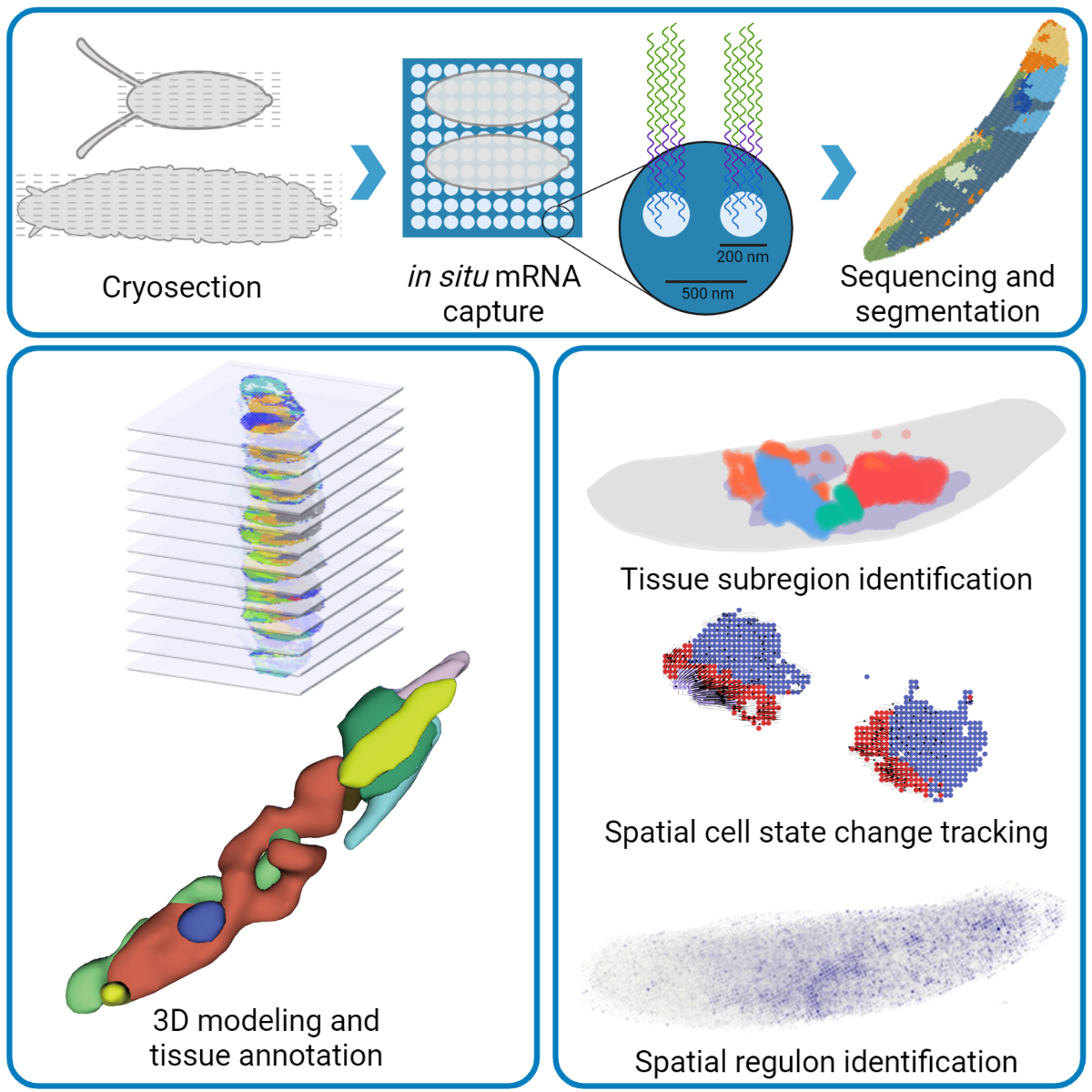
Figure 4. Experimental procedures of this study
Dr. Mingyue Wang, joint postdoctoral fellow between the School of Life Sciences at SUSTech and BGI-Shenzhen, Dr. Qinan Hu, postdoctoral fellow from the School of Medicine at SUSTech, Dr. Qing Lan, Associate Researcher at BGI Shenzhen, along with graduate students from BGI-Shenzhen Tianhang Lv, Yuhang Wang, Rong Xiang, and Zhencheng Tu, are the co-first authors of this paper.
Dr. Longqi Liu, Director of BGI-Research Cell Science Institute, Prof. Yuhui Hu of the School of Medicine at SUSTech, Dr. Xun Xu, Director of BGI Shenzhen, Dr. Xiaoshan Wang, Associate Researcher at BGI-Shenzhen, and Dr. Yuxiang Li, Chief Scientist of Bioinformatics at BGI-Research, are the corresponding authors.
This work was supported by the Shenzhen Key Laboratory of Gene Regulation and Systems Biology, Shenzhen Science and Technology Program, National Natural Science Foundation of China (NSFC), and the Presidential Postdoctoral Fellowship.
This work is also part of the initial publication release of STOC (SpatioTemporal Omics Consortium). STOC is a newly founded global collaborative research initiative that aims to accelerate our understanding of cellular complexity and interactions at the tissue scale in development, physiology, and disease through large-scale spatially resolved multi-omics analyses.
As one of the initial members of STOC, Prof. Yuhui Hu’s lab in the School of Medicine at SUSTech focuses on system biology and functional genomics. It has been developing system biology methods in disease progression and drug mechanisms involving processes such as tumor, development, and metabolism, with a focus on frontier techniques including high-throughput sequencing, single-cell multi-omics, spatial transcriptomics, CRISPR-mediated genome editing, proteomics, and metabolomics.
Paper link: https://doi.org/10.1016/j.devcel.2022.04.006
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By