The hippocampus is an essential structure in the brain responsible for encoding and storing memories. The mammalian hippocampus consists of three major subregions: DG (dentate gyrus), CA3, and CA1. These three subregions are interconnected and form neural networks with cortical neurons to support the passage and storage of information.
N6-methyladenosine (m6A), the most abundant RNA modification, regulates RNA fate at the epitranscriptome level and modulates nervous system development and function through its reader proteins. Though it is known that m6A is crucial for memory formation, which reader is involved in the memory storage related to the hippocampus and which subregion of the hippocampus plays an important role in this process, remains to be studied.
Associate Professor Sheng-Jian Ji’s research group from the Department of Neuroscience, School of Life Sciences, at the Southern University of Science and Technology (SUSTech) recently published a research article that found that the reader protein YTHDF2 of m6A modification plays an important role in mouse hippocampus-dependent learning and memory.
Their study, entitled “YTHDF2 in the dentate gyrus is the m6A reader mediating m6A modification in hippocampus-dependent learning and memory,” was published in the academic journal Molecular Psychiatry.
Prof. Ji’s Lab generated DG, CA3, and CA1 subregion-specific Ythdf1 and Ythdf2 knockout mice, respectively. Unlike previous reports, they found that conditional knockout of Ythdf1 from three subregions of the hippocampus did not impair spatial learning and memory, and only DG-specific but not CA3- or CA1-specific Ythdf2 knockout mice showed memory deficits.
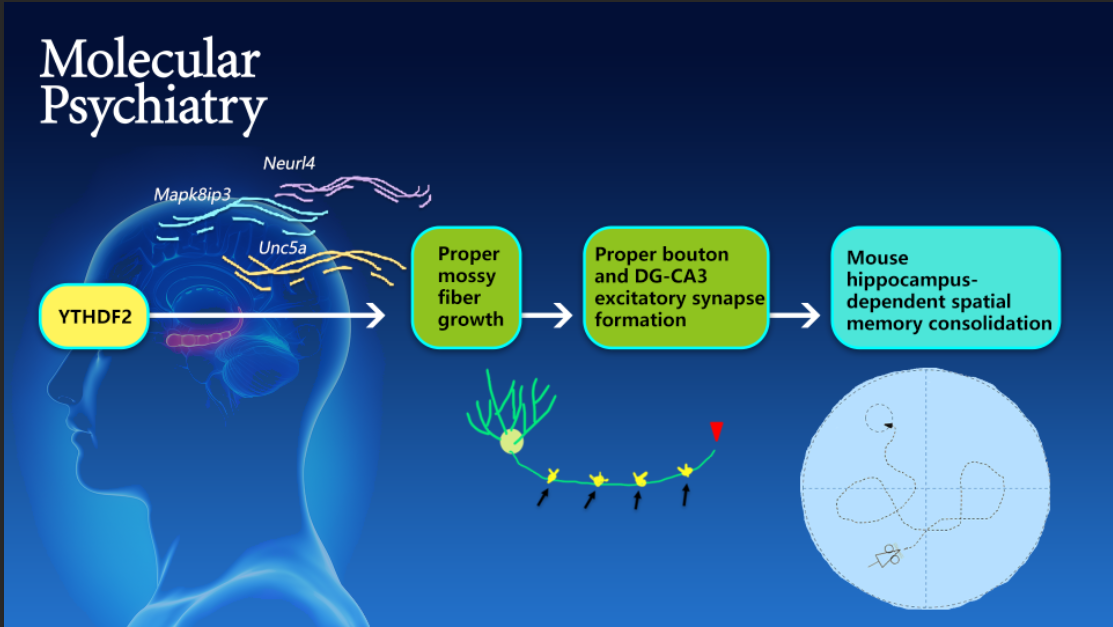
To explore the regulatory mechanism, they first studied the neurogenesis of granule cells in the DG subregion as well as the development of their axons and dendrites. They found that the loss of Ythdf2 in the DG caused the overgrowth of mossy fibers in the axons of granule cells, which further led to defects of the excitatory synapse formation and functional impairment on DG-CA3 synapses. By combining RNA-seq technology and in vitro/in vivo experiments, they established the regulation model that YTHDF2 in DG regulates the growth of mossy fibers by controlling the stability of its m6A-modified target mRNAs, which encode proteins that promote axon growth and elongation.
This study proves that YTHDF2 in DG is the only m6A reader that mediates m6A modification in regulating memory formation, supplementing and expanding our understanding of the roles of m6A modification on memory formation in the field. This work was highly commended by the reviewers, who commented that “This (study) overturns the dogma in the field” and “This study will no doubt be a landmark in the field”.
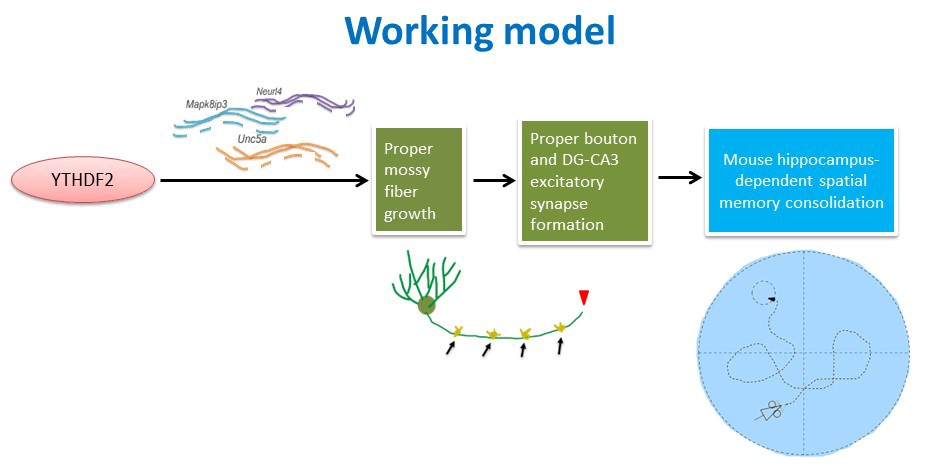
Figure 1. Working model
Mengru Zhuang, a SUSTech-HKUST joint Ph.D. program candidate from Assoc. Prof. Sheng-Jian Ji’s Lab (currently a postdoctoral fellow at SUSTech), Xiaoqi Geng, Lecturer of Neurosurgery at the Affiliated Hospital of Southwest Medical University (SMU), Peng Han, a former Research Associate at Prof. Ji’s Lab, and master’s graduates Pengfei Che (SUSTech), Fanghao Liang (SUSTech) and Chao Liu (SMU) are the co-first authors of this paper. SUSTech is the primary affiliated institute. Assoc. Prof. Sheng-Jian Ji and Wei Dong, a PI at the Key Laboratory of Medical Electrophysiology of the Ministry of Education, SMU, are the co-corresponding authors.
This study was supported by the National Natural Science Foundation of China (NSFC), Shenzhen-Hong Kong Brain Science Innovation Institute, and Shenzhen Key Laboratory of Gene Regulation and Systems Biology.
It is worth noting that the Ji Lab has published six research articles in the field of m6A modification regulating neural development and functions, and has become (one of) the most important laboratories in this field in the world.
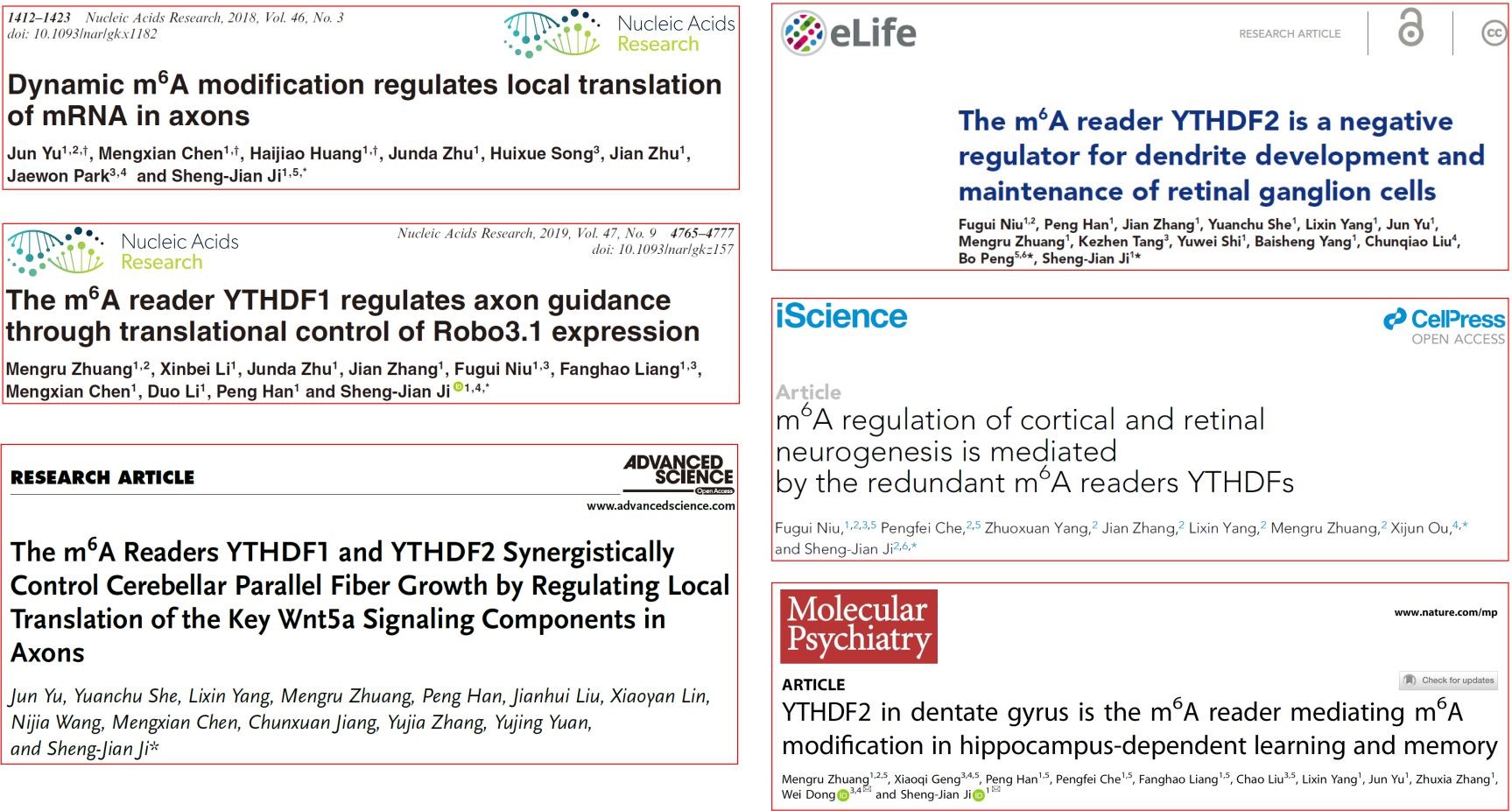
Figure 2. The research articles published by the Ji Lab in the field of m6A modification regulating neural development and functions
Paper link: https://www.nature.com/articles/s41380-023-01953-z
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By