The CRISPR-Cas system represents a widespread adaptive immune mechanism in bacteria and archaea, capable of recognizing and cleaving foreign genetic materials such as phage DNA or RNA. To counteract this immune defense, phages have evolved diverse anti-CRISPR systems.
A research team led by Associate Professor Ning Jia from the Department of Biochemistry at the Southern University of Science and Technology (SUSTech), in collaboration with Professor Alexander J. Meeske’s team from the University of Washington, has identified a novel endogenous small RNA molecule named rAcrVIA1 that inhibits CRISPR-Cas13 activity by structurally mimicking crRNA. This mimicry enables foreign nucleic acids to evade host immune defenses. This discovery of new RNA-type anti-CRISPR molecules (rAcrs) expands our understanding of CRISPR regulatory mechanisms and provides novel insights for developing RNA-based precision CRISPR modulation tools.
Their paper, titled “RNA-mediated CRISPR-Cas13 inhibition via crRNA structural mimicry”, has been published in Science.
The Ning Jia research group has long focused on uncovering the molecular mechanisms by which prokaryotic immune systems, including those in bacteria and archaea, defend against phage and plasmid invasions. In recent years, they have revealed the molecular mechanisms of various prokaryotic immune systems, including CRISPR and pAgos systems, in defending against phage and plasmid attacks (Nature Chemical Biology, 2024; Molecular Cell, 2023, 2019a, 2019b, 2019c; Nature Communications, 2024a, 2024b, 2022; Science, 2020; Cell Research, 2024, 2022, 2020; Nature Reviews Molecular Cell Biology, 2021, etc.). While most reported anti-CRISPR systems are protein-encoded, with some RNA-based anti-CRISPRs sharing high sequence similarity with crRNAs and functioning through crRNA displacement, it remained unclear whether other types of anti-CRISPR systems exist or whether RNAs without sequence similarity to crRNAs could also exert inhibitory effects.
Through experimental investigations, the researchers identified a small non-coding sequence upstream of a known anti-CRISPR protein that directly inhibits the CRISPR-Cas13 system (Figure 1). Small RNA sequencing confirmed that this sequence is transcribed into a 37-60 nt non-coding RNA. Structural predictions indicated that this RNA forms two stem-loop structures, and subsequent mutagenesis experiments demonstrated that these secondary structures are crucial for its inhibitory function, leading to its designation as rAcrVIA1.

Figure 1. A small non-coding RNA (rAcrVIA1) inhibits type VI CRISPR-Cas system activity
Co-expression and pull-down experiments demonstrated that Cas13 forms a stable complex with rAcrVIA1. However, mutations in key residues of Cas13 involved in crRNA maturation abolished complex formation, suggesting that rAcrVIA1 and crRNA are processed by Cas13 in a similar manner. Cryo-EM structural analysis of the Cas13-rAcrVIA1 complex revealed that, despite lacking sequence similarity, rAcrVIA1 adopts a similar overall structure to crRNA (Figure 2). This structural mimicry enables rAcrVIA1 to inhibit Cas13 activity. Unlike crRNA, rAcrVIA1 cannot bind target RNA, thereby locking Cas13’s HEPN active sites in an inactive state.
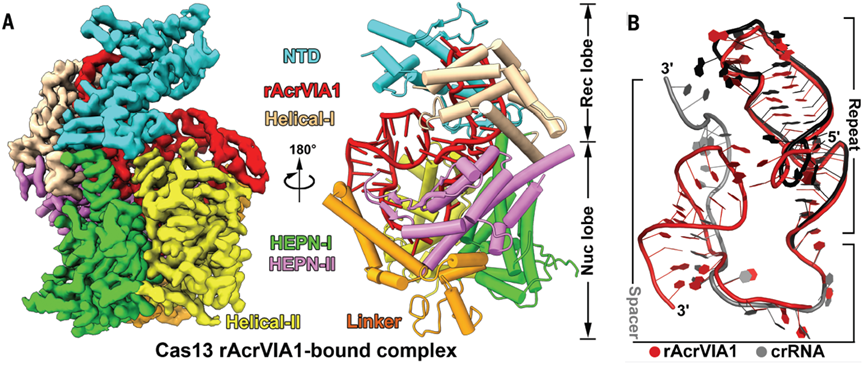
Figure 2. rAcrVIA1 and crRNA fold into similar overall structures
The study also found that rAcrVIA1 coexists with another Cas13 inhibitor protein, AcrVIA2, within the same gene cluster. These two inhibitors employ distinct mechanisms: rAcrVIA1 competitively inhibits Cas13-crRNA binding through structural mimicry, while AcrVIA2 degrades crRNA. This dual inhibition mechanism likely enhances immune evasion during mobile genetic element invasion.
Additionally, the researchers discovered that rAcrVIA1 expression is regulated by the upstream gene orf1, which encodes a helix-turn-helix (HTH) domain-containing protein. ORF1 acts as a transcriptional autorepressor, fine-tuning rAcrVIA1 production. Loss of ORF1 suppresses rAcrVIA1 expression, rendering it unable to effectively inhibit Cas13. This finding unveils the sophisticated regulatory mechanism governing rAcrVIA1 under natural conditions.
This study not only uncovers a novel RNA anti-CRISPR mechanism but also opens new avenues for developing RNA-based CRISPR modulation tools. By mimicking crRNA structure, rAcrVIA1 effectively inhibits Cas13 activity, offering exciting possibilities for designing more precise gene-editing technologies.
Jun-Tao Zhang from SUSTech and Victoria M. Hayes from the University of Washington are the co-first authors of this paper. Associate Professor Ning Jia and Professor Alexander J. Meeske serve as the co-corresponding authors.
Paper link: http://doi.org/10.1126/science.adr3656
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yuwen ZENG
Photo ByDepartment of Biochemistry