As information technology advances, ensuring data security and privacy protection has become increasingly challenges. DNA has emerged as a promising medium for information storage due to its high density and durability. However, ensuring the secure transmission of DNA-encoded information to prevent unauthorized access remains a significant scientific hurdle.
Conventional DNA sequencing technologies and their associated basecallers are primarily designed for natural DNA and often struggle to accurately interpret chemically modified DNA. This limitation presents both a challenge and an opportunity for developing DNA-based private communication. Yet, the difficulty in training basecallers for such modified DNA has impeded its practical application.

To address this, a research team led by Associate Professor Yi Li from the School of Microelectronics at the Southern University of Science and Technology (SUSTech) has made notable advancements in DNA information storage and private communication by proposing DeepSME, a framework leveraging nanopore sequencing and deep learning.
Their study, titled “De novo non-canonical nanopore basecalling enables private communication using heavily-modified DNA data at single-molecule level”, has been published in the international academic journal Nature Communications.
The DeepSME framework facilitates the “de novo” construction of a basecaller specifically for DNA that has undergone heavy chemical modification, such as the replacement of all natural cytosine (C) bases with 5-hydroxymethylcytosine (5hmC). This non-natural chemical modification substantially disrupts the readout by conventional basecallers, leading to high error rates or failure to decode, thereby “hiding” the information and enhancing communication privacy (Figure 1). DeepSME acts as a corresponding “key” to accurately decrypt this concealed molecular information.
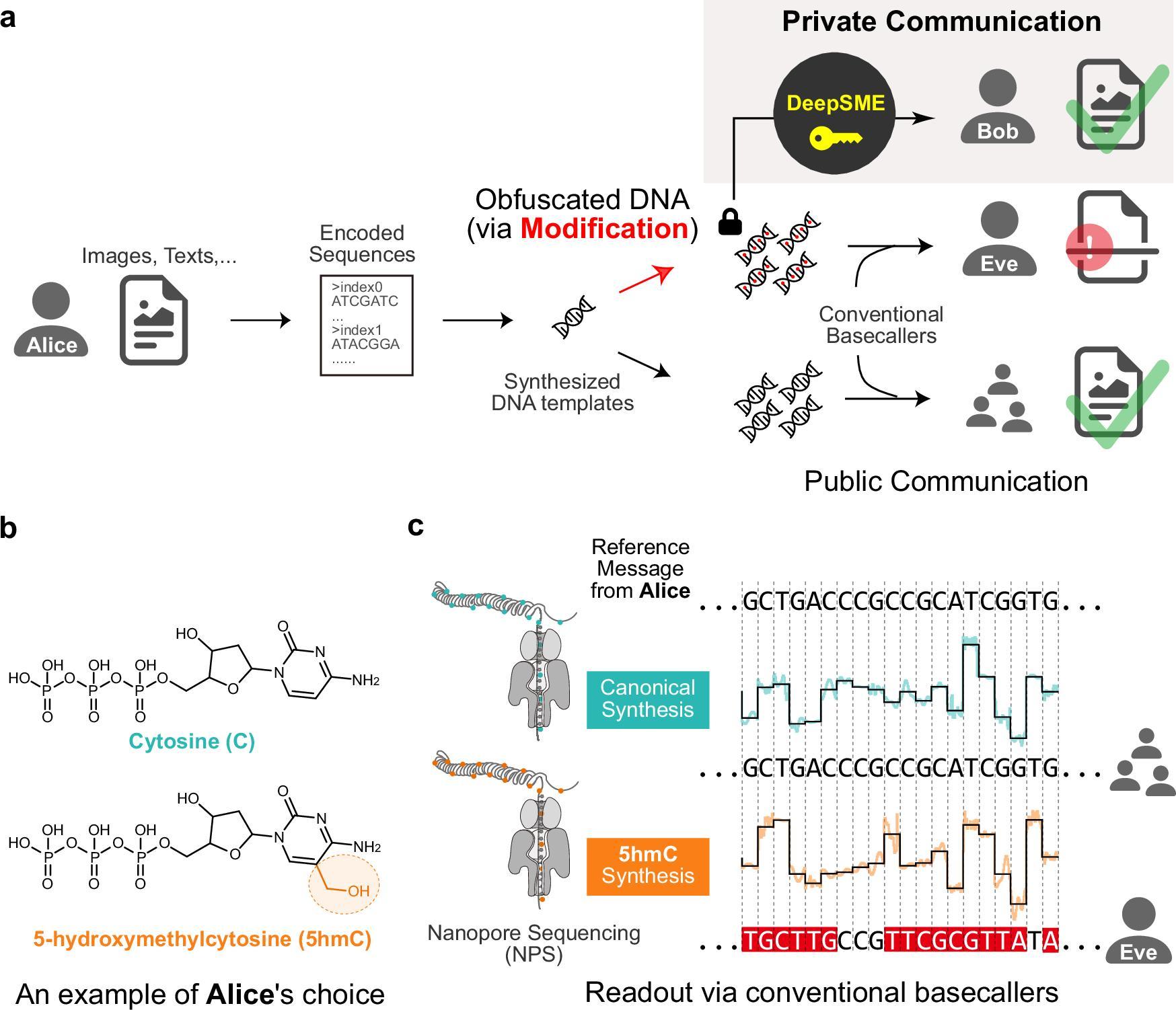
Figure 1. Scheme of private communication using modified DNA
The researchers developed an innovative three-stage, alignment-free training pipeline for DeepSME (Figure 2a-b). This approach generates a k-mer (short DNA sequence fragments) dictionary from scratch, addressing the challenges of processing modified DNA, which lacks reliable alignment references and pre-existing models. The pipeline then utilizes simulated sequence current data for initial enhancement, followed by real sequencing data from biological samples for reinforcement.
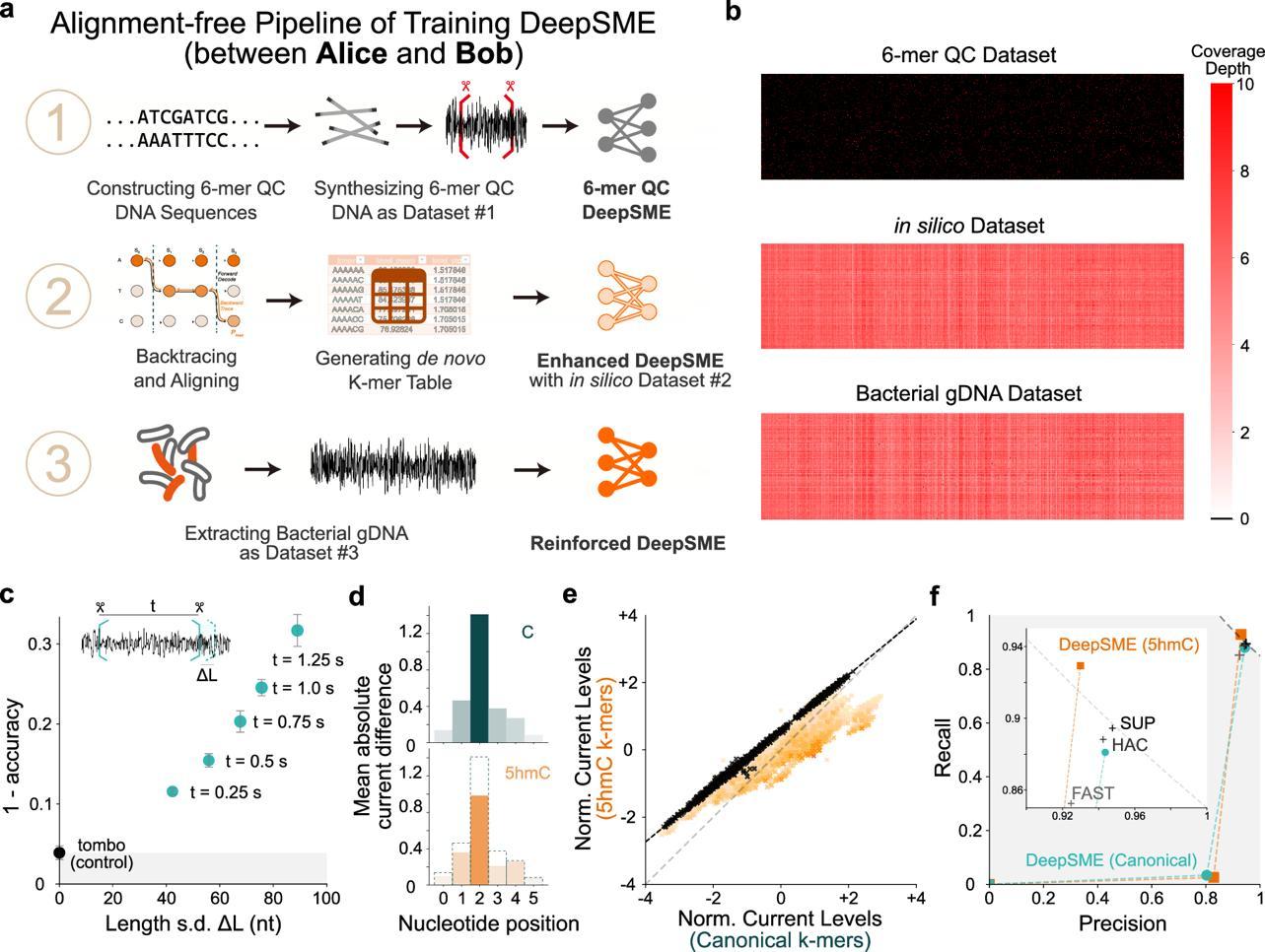
Figure 2. Three step training pipeline and the basecalling performance of DeepSME
The resulting DeepSME basecaller demonstrated strong performance, achieving over 92% precision and recall (Figure 2f) and an F1-score of 86.4%. These metrics surpassed that of current state-of-the-art commercial and open-source basecallers when applied to the heavily-modified DNA (Figure 3f).
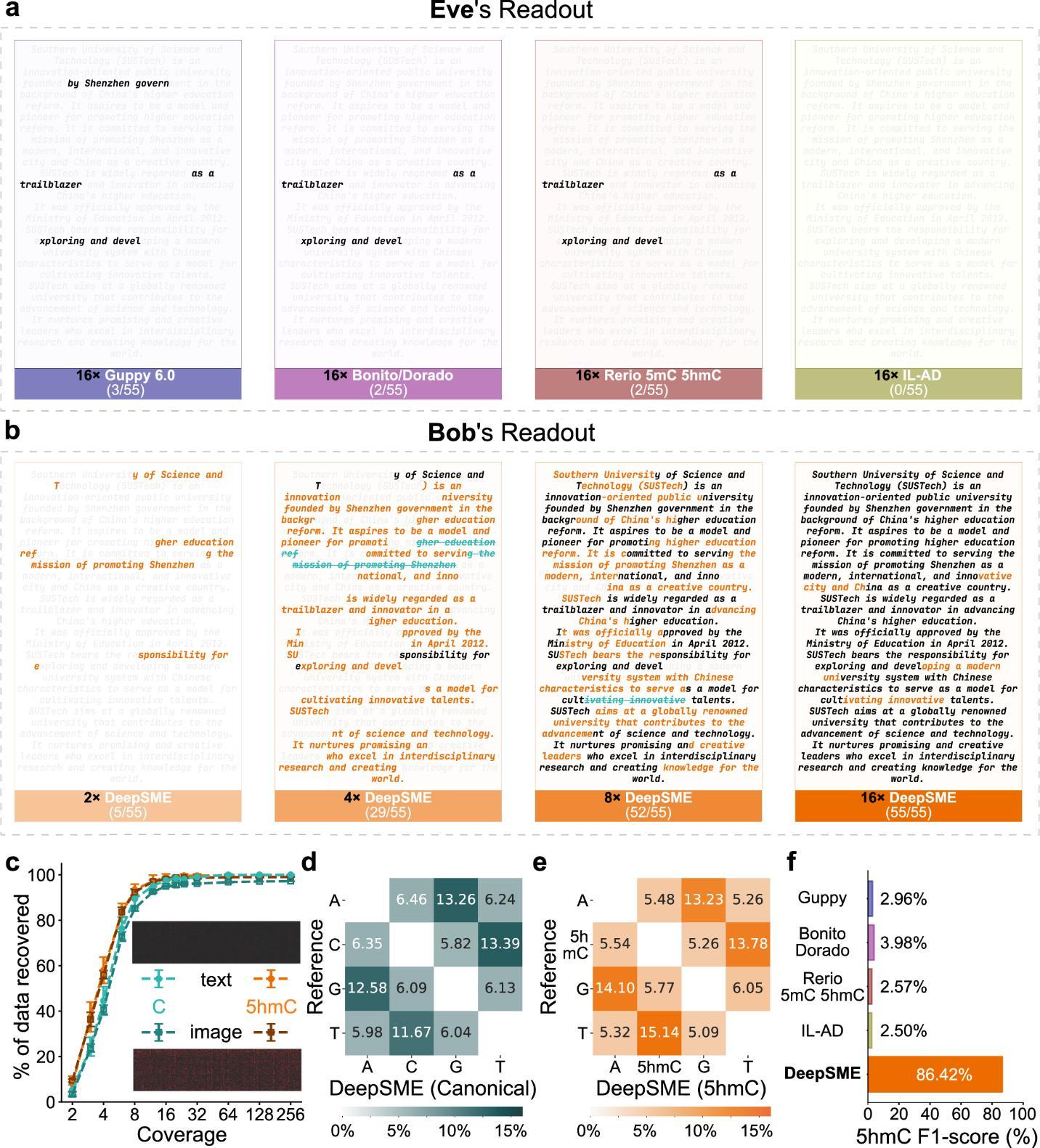
Figure 3. Performance on decoding concealed information from modified DNA with DeepSME
Experimental results showed that DeepSME, when combined with the team’s Composite Hedges Nanopores (CHN) DNA encoding scheme (see related link below), successfully decrypted text and image files encoded in fully 5hmC-modified DNA. For an unsuspecting third party (Eve) using standard basecallers, virtually no valid information could be recovered (Figure 3a). Conversely, a recipient (Bob) possessing the DeepSME “key” could efficiently and accurately retrieve the complete original text information with only 16× sequencing coverage (Figure 3b). These findings demonstrate the potential of the DeepSME framework for private communication using DNA.
This research offers a novel approach for privacy in DNA data storage and transmission and highlights the capability of deep learning in interpreting complex biomolecular signals. The DeepSME framework, with its alignment-free training pipeline, potential for customization, efficient training procedure, and relatively moderate computational demands, may find broader applications in bioengineering, information security, anti-counterfeiting, and medical diagnostics.
Master’s student Qingyuan Fan from the School of Microelectronics is the first author of the paper. Associate Professor Yi Li is the corresponding author, with SUSTech as the first affiliated institution.
Paper link: https://doi.org/10.1038/s41467-025-59357-2
Related link: https://doi.org/10.1038/s41467-024-53455-3
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yuwen ZENG
Photo BySchool of Microelectronics