The circadian clock serves as an endogenous time-keeping mechanism in plants, enabling them to perceive periodic changes in the external environment and execute key physiological functions at appropriate times. Recent studies have shown that the plant circadian clock exhibits tissue and cell-type specificity in both its molecular components and functional regulation. This heterogeneity allows plants to perceive and respond to environmental changes more precisely, optimizing growth and survival strategies. However, traditional research methods, such as whole-tissue sequencing or luciferase reporter assays, are inefficient for resolving differences in rhythmic gene expression at the single-cell level. In addition, the presence of plant cell walls adds significant technical difficulty to single-cell isolation.
A research group led by Professor Jixian Zhai from the Department of Biology, School of Life Sciences at the Southern University of Science and Technology (SUSTech) has now constructed a circadian gene expression atlas at single-cell resolution using single-nucleus RNA sequencing (snRNA-seq) technology. Their work reveals cell-type-specific circadian gene expression patterns in Arabidopsis thaliana, identifying a novel circadian regulator.
Their paper, titled “48-Hour and 24-Hour Time-lapse Single-nucleus Transcriptomics Reveal Cell-type specific Circadian Rhythms in Arabidopsis”, has been published in Nature Communications.
To gain a comprehensive understanding of circadian gene expression patterns at the single-cell level, the research team employed single-nucleus RNA sequencing (snRNA-seq) to Arabidopsis seedlings under continuous light. They performed two time-series sampling regimes: a high-resolution “dense” sampling every two hours over 24 hours, and a “long-duration” sampling every four hours extended to 48 hours. Using the 10x Genomics platform, they measured the transcriptomes of over 200,000 nuclei, constructed a single-cell transcriptional atlas of the Arabidopsis circadian rhythm, and dissected the expression differences of core clock genes across distinct cell types (Figure 1).
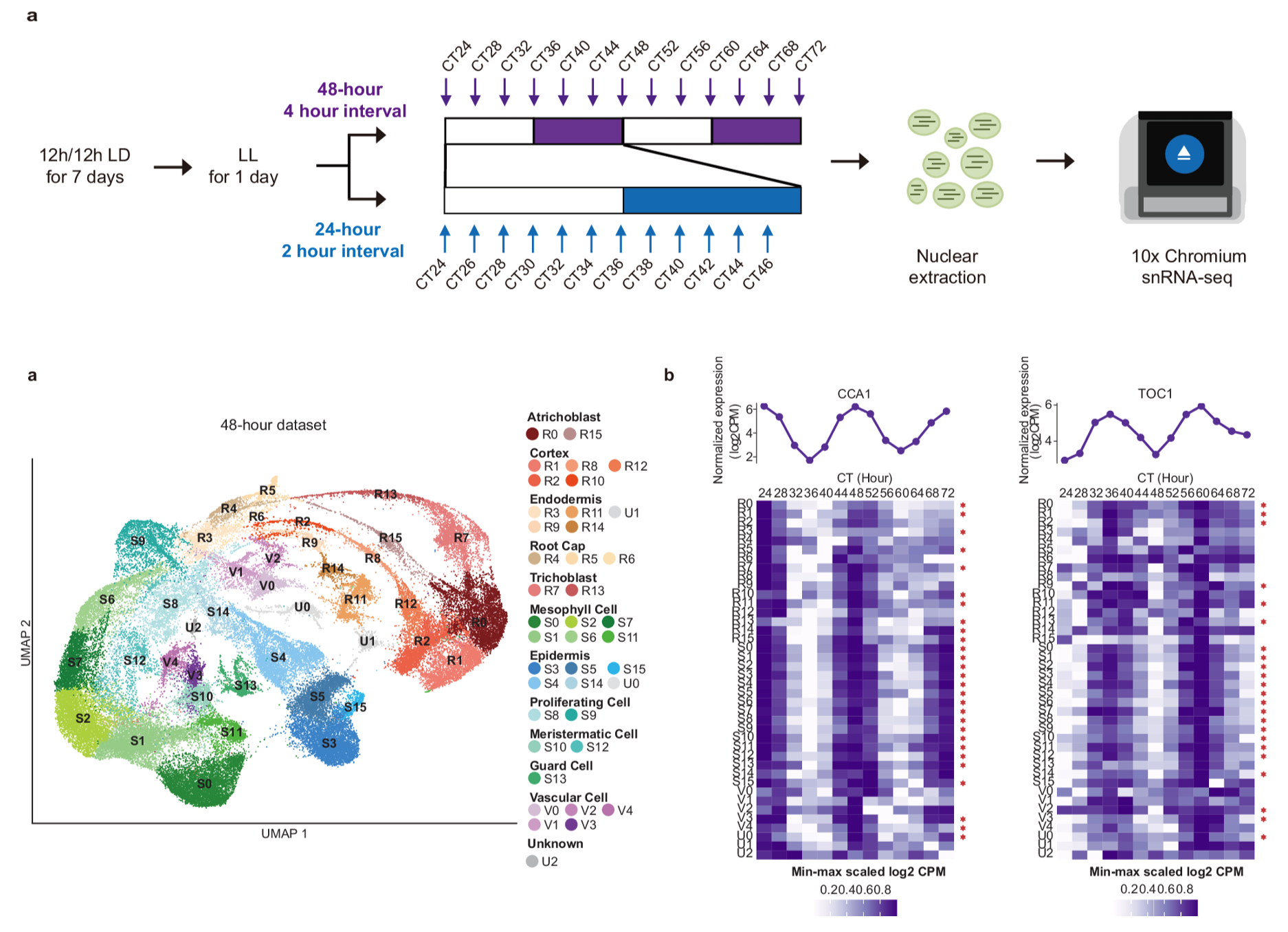
Figure 1. Construction of the Arabidopsis circadian single-cell transcriptional atlas using single-nucleus sequencing.
Their analysis showed that the majority of oscillating genes were specific to particular cell types, with significant differences observed between shoot and root. Furthermore, comparisons of oscillating genes across different cell clusters revealed that the sets of oscillating genes in four types of mesophyll cells (S0, S1, S2, S7) were highly similar (Figure 2). Pairwise phase analysis of these shared oscillating genes found that the expression phases of most genes were delayed in S0 and S1 compared to S2 and S7 (Figure 2), suggesting the existence of a more refined circadian clock regulatory mechanism within mesophyll cells.
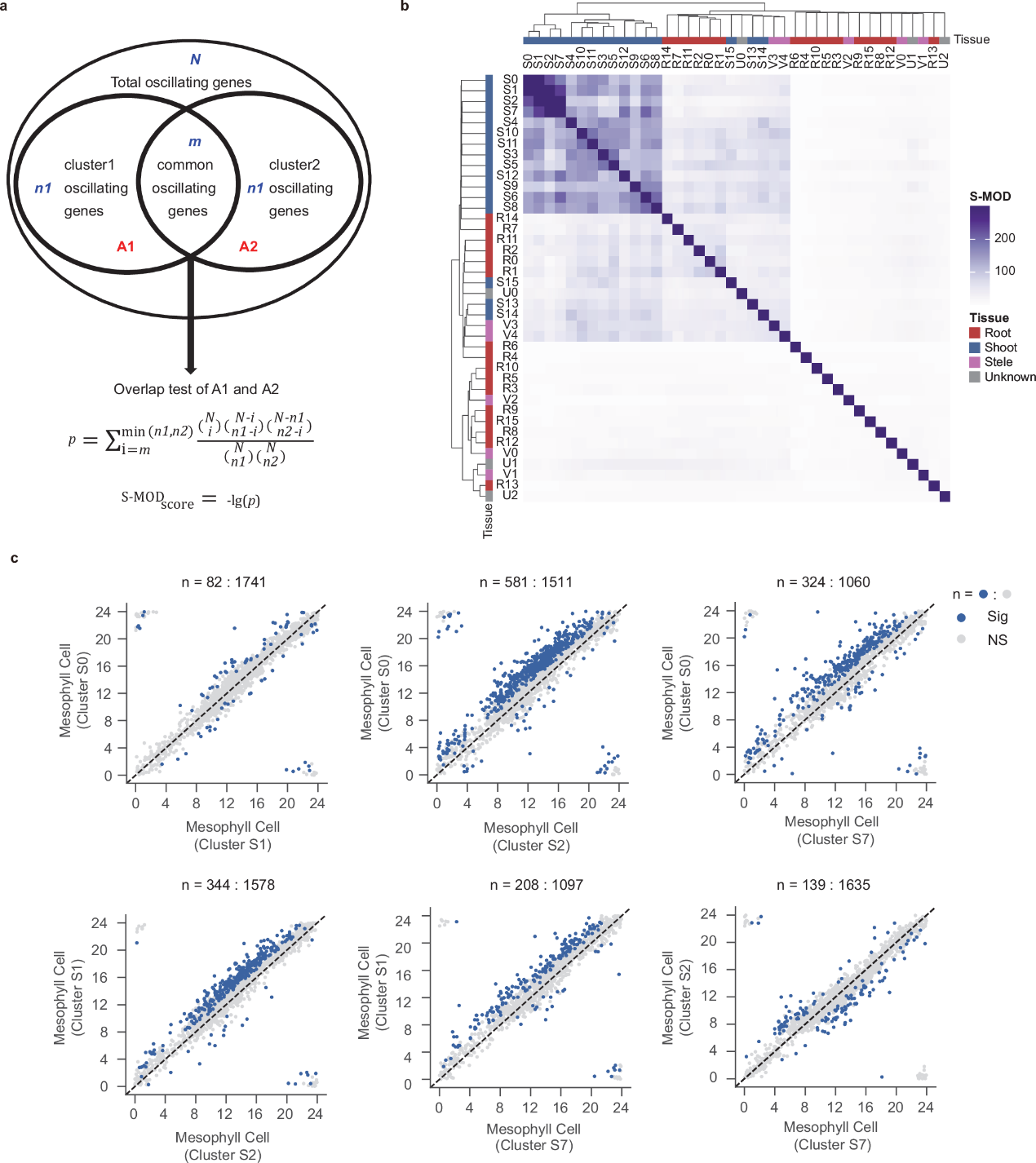
Figure 2. Comparative analysis of oscillating gene sets between different cell types
The researchers revealed that genes encoding circadian regulators oscillated across multiple cell types, with the majority being known core clock genes (Figure 3). Notably, ABF1 was identified as a novel circadian regulator whose overexpression significantly shortened the circadian period (Figure 3). These findings further refine the gene regulatory network of the plant circadian clock and provide new molecular entry points for systematically analyzing the mechanisms of rhythmic oscillation.
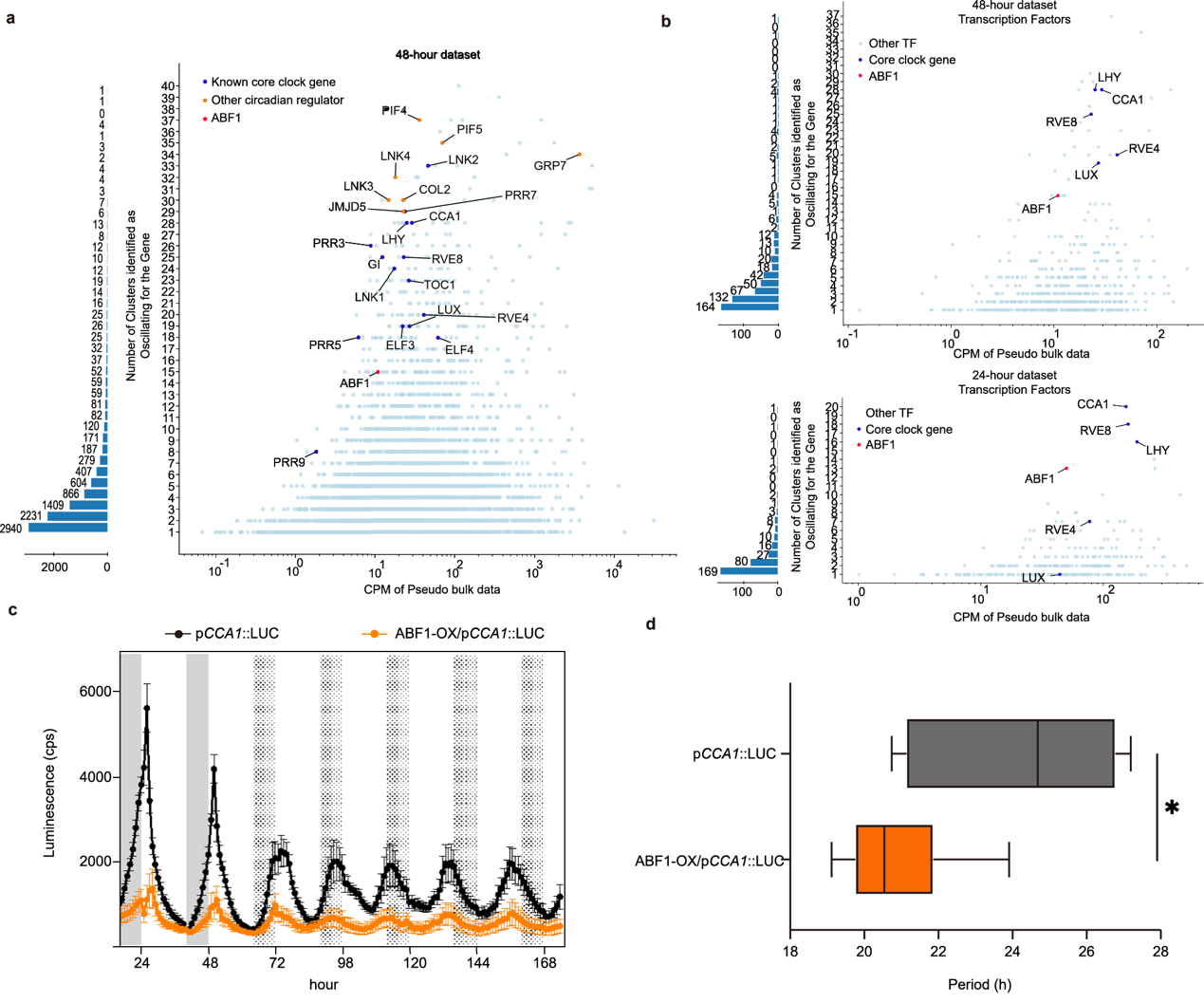
Figure 3. Identification of circadian regulators
The Arabidopsis circadian single-cell atlas established in this study has been integrated into a data resource website (see link below). It will provide crucial data support for subsequent research in the circadian rhythm field (Figure 4).
Figure 4. Website homepage
Dr. Yuwei Qin and Research Associate Professor Yanping Long from SUSTech, together with Researcher Zhijian Liu from Northeast Normal University, and Ph.D. students Shiqi Gao and Carlos Martínez-Vasallo from Henan University and IBMCP, Universitat Politècnica de València, respectively, are the co-first authors of the paper. Professor Qiguang Xie from Henan University, Professor María A. Nohales from IBMCP, Universitat Politècnica de València, and Professor Jixian Zhai are the corresponding authors.
Other contributors to this work include Professor Xiaodong Xu and Ya Gao from Henan University, Researcher Bin Liu from the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, and Ph.D. student Xinlong Zhu from SUSTech.
Paper link: https://doi.org/10.1038/s41467-025-59424-8
Circadian data resource website: https://zhailab.bio.sustech.edu.cn/sc_circadian
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yuwen ZENG
Photo BySchool of Life Sciences