With global food security an on-going issue for the world, molecular biologists around the world have been working on how to understand the operation of plants better. A group of Shenzhen scientists has discovered essential features about how plants decode genetic information from DNA to RNA, a fundamental process within a cell that instruct the activity of life on Earth.
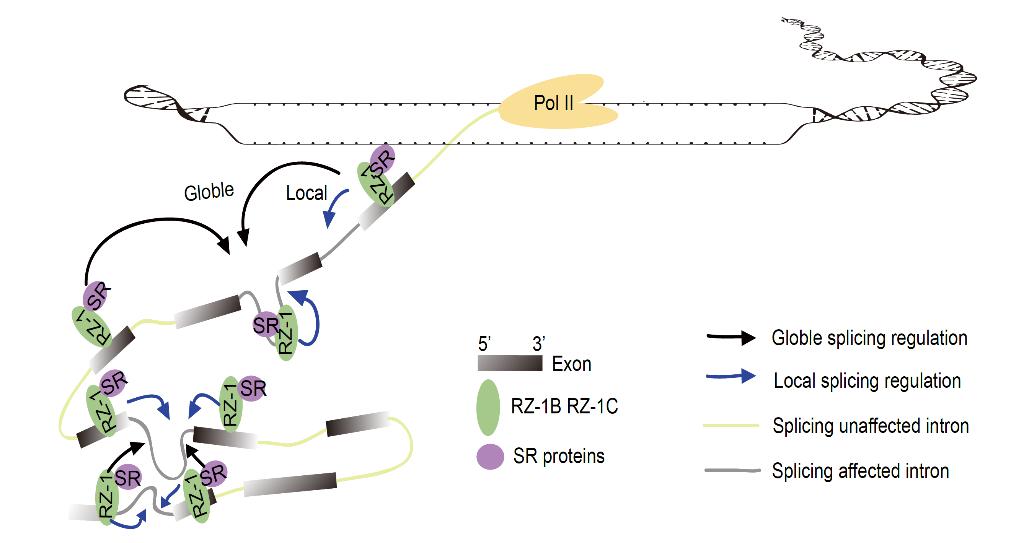
Figure 1: Efficient co-transcriptional splicing likely being promoted through high-dimensional interactions between introns, exons, and splicing factors.
A research group from the Department of Biology at Southern University of Science and Technology (SUSTech) and the SUSTech-Peking University (PKU) Institute of Plant and Food Science have worked together to publish a paper online with Molecular Plant (IF = 10.8). The paper is titled, “The features and regulation of co-transcriptional splicing in Arabidopsis.” The paper systematically studied the coupling between RNA splicing and transcription, revealing the regulatory mechanisms behind RNA splicing at chromatin levels in plants.
The research team focused on a fundamental process called transcription. In this process, DNA is used as a template to instruct the synthesis of RNA (transcription). The newly synthesized RNA may contain parts (intron) that need to be removed before such RNA can be used as a template to synthesize proteins. The removal of introns and re-joining together the essential parts (exons) are known as RNA splicing.
In the past 30 years, people have had a deep understanding of the mechanism of RNA splicing in yeast and animal cells, but the understanding of the mechanism of plant RNA splicing is still limited. Although the structure and function of the splicing complex (spliceosome) are conserved in yeast and humans, there may still be significant differences in the splicing regulatory mechanisms of different species.
The research team used Arabidopsis thaliana, a plant that has been used by thousands of research teams around the world as a model plant. The team discovered that RNA splicing in plants is, in general, happening during the process of transcription, a process known as co-transcriptional splicing, which is also the case in a different model organism such as yeast, insect and human cells. They found that rapid transcription is unfavorable for co-transcriptional splicing, consistent with the kinetic coupling model raised based on data in non-plant species. They also found similar patterns of alternative splicing between mRNA and chromatin-bound RNA, suggesting alternative splicing events are mainly determined at the chromatin level. The team also discovered, for the first time, that co-transcriptional splicing efficiency is correlated with the number of introns, but not with gene length.
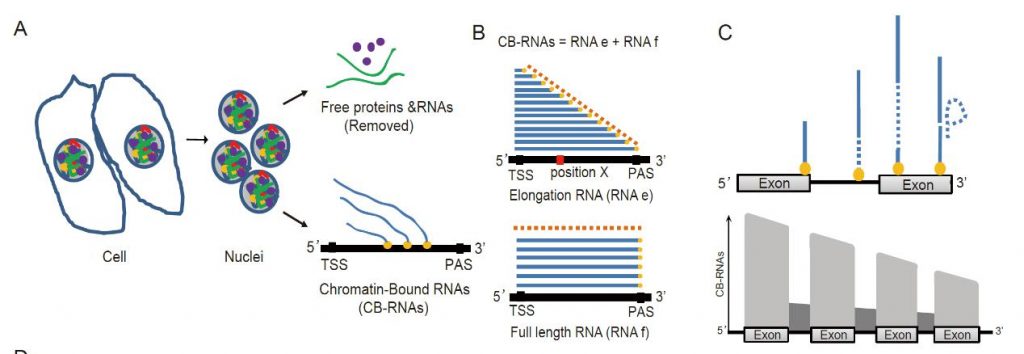
Figure 2: A: Schematic showing the source and extraction process of CB-RNA. B: CB-RNA corresponds to two fractions of RNA, where RNAe represents the RNA that is being transcribed and extended by RNA polymerase. RNAf represents RNA whose transcriptional extension has almost ended but has not yet been detached from chromatin. C: Assuming that the RNA is co-transcriptionally spliced, the expected distribution of CB-RNA along a gene is shown.
One vital area is the regulatory mechanisms behind co-transcriptional splicing mediated by proteins. The SUSTech researchers found that exons and introns can be used as binding sites of a splicing factor (RNA-binding proteins) called RZ-1, which promote the efficient co-transcriptional RNA splicing. They further propose that the higher-order interactions between exons, introns, and splicing factors could be critical for efficient co-transcriptional splicing.
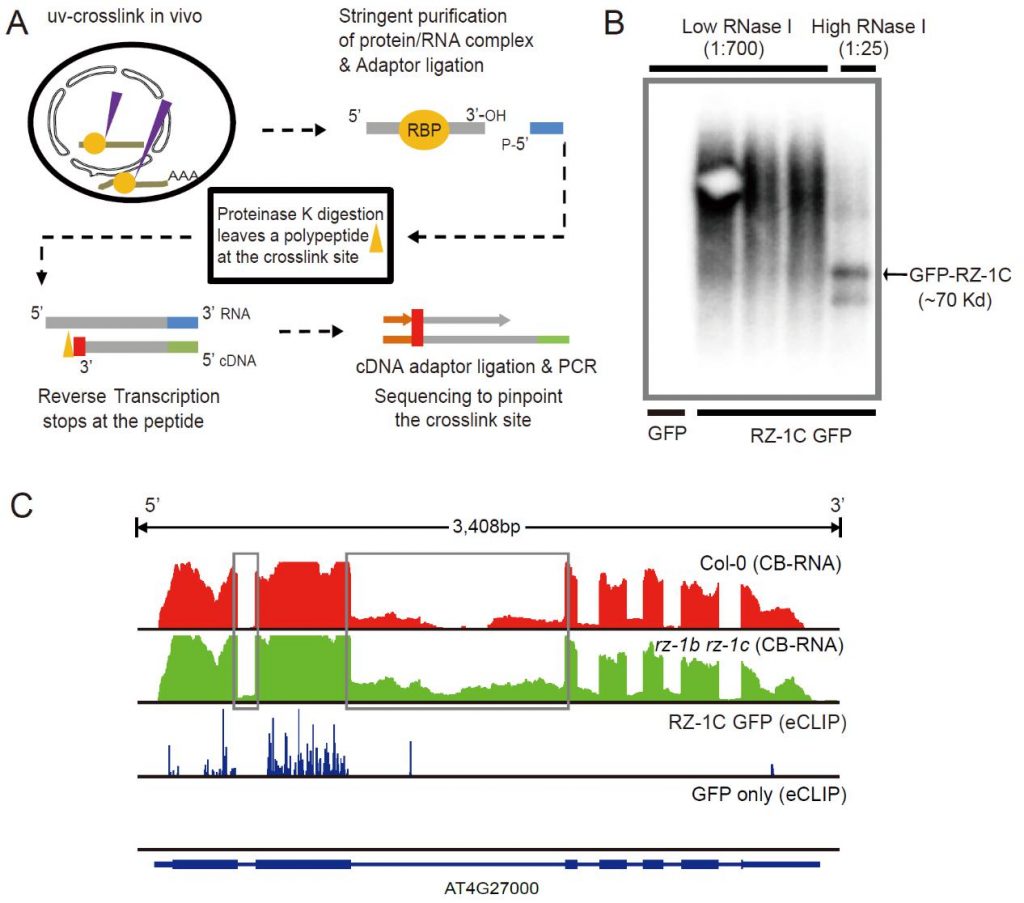
Figure 3: A: A schematic diagram showing the principle and experimental flow of eCLIP-seq. B: Autoradiographic image of RNA bound by RZ-1C protein captured in eCLIP-seq experiments. C: An example of a gene that is directly regulated by RZ-1C in a co-transcriptional scissor. The picture shows the CB-RNA-seq results and eCLIP-seq results in the gene.
As part of their research, the authors adapted two vital experimental technologies for plant research. The first technique (CB-RNA-seq) can be used to study the status of transcribing (chromatin-bound) RNA. In combination with a simple mathematical model, the distribution of chromatin-bound RNA along the chromatin can be used to study the initiation, elongation, and termination (chromatin RNA release) rate of transcription. The second technique (eCLIP-seq) locates RNA-binding protein binding substrates & binding sites with much higher accuracy and efficiency than previous technology. Both technological developments will accelerate RNA and transcription-related research in plants.
Professor Wu Zhe’s research group led the research, in conjunction with Professor Wu Yufeng’s research group from the Bioinformatics Center at Nanjing Agriculture University. SUSTech Research Assistant Professor Zhu Danling and Nanjing Agriculture University doctoral candidate Mao Fei were the co-first authors. Professor Wu Zhe and Professor Wu Yufeng were the correspondent authors. The project was initiated under the support and inspiration from Professors Qu Li-Jia and Gu Hongya from the Peking-Tsinghua Center for Life Sciences at Peking University and Professor Dame Caroline Dean from the John Innes Centre, UK. Professor Gu Lianfeng from the College of Forestry at Fujian Agriculture & Forestry University was also involved in the research.
This project was supported by the start-up funding of SUSTech, the National Natural Science Foundation of China, the Science and Technology Department of Guangdong Province, and the Shenzhen Municipal Science and Technology Commission.
Article link: https://www.sciencedirect.com/science/article/pii/S1674205219303685
Proofread ByXia Yingying
Photo ByDepartment of Biology, Institute of Plant and Food Science