The abnormality of tubulin tyrosination or detyrosination levels are associated with severe human pathologies including cancer, heart failures, cardiomyopathies, and brain dysfunctions. The tubulin detyrosination-tyrosination cycle is initiated by tubulin tyrosine carboxypeptidase (TCP), which is comprised of the vasohibin-SVBP enzyme complex in human. The detailed understanding of the catalytic mechanism of vasohibin-SVBP is important to improve the drug development opportunities toward this unique enzyme.
Last month, research led by Associate Professor Huang Hongda from the Department of Biology at Southern University of Science and Technology (SUSTech) was published in Nature Structural and Molecular Biology. The paper, entitled “Structural basis of tubeulin in detyrosination by the vasohibin-SVBP enzyme complex,” elucidated the molecular mechanism of tubulin detyrosination by the VASH2-SVBP protein complex.
Previous research had indicated that the tubulin detyrosination-tyrosination cycle played an important role in maintaining the functioning of microtubules, the microscopic tubular structures present in the cytoplasm of cells. It has also been found that detyrosination level of tubulin in stable microtubules was higher than in dynamic microtubules. An imbalance in the tubulin detyrosination-tyrosination cycle leads to dysfunction in the microtubules and could lead to serious illness. However, it was not previously known how the vasohibin-SVBP enzyme complex catalyzes the detyrosination of tubulin.
The research paper analyzed the crystal structures of the VASH2-SVBP enzyme complex. Their experiments showed that SVBP can correctly fold VASH2 while also playing an important role in detyrosinating the tubulin. Their research also discovered the mechanism of two inhibitors.
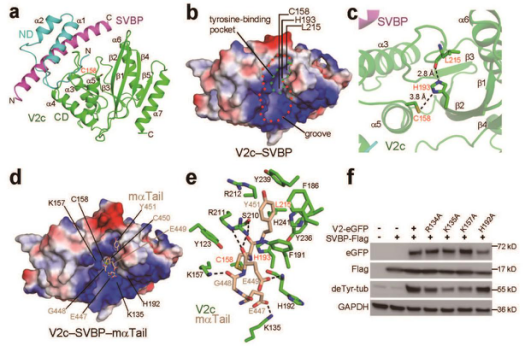
The researchers have ultimately found the structural basis for the detyrosination of tubulin by VASH2-SVBP, and its role in neuron development. Their discovery paves way for future research of small molecule inhibitors, along with a unique platform for developing drugs against human conditions with abnormal tubulin tyrosination levels, such as cancer, heart defects and possibly brain disorders.
This specific paper was a multinational effort. Associate Professor Huang Hongda worked with Dr. Marie-Jo Moutin from the Grenoble Institut Neurosciences at the University Grenoble Alpes in France and Professor Michel. O Steinmetz of the Laboratory of Biomolecular Research at the Paul Scherrer Institute in Switzerland. The Department of Biology at SUSTech was the first communications unit. SUSTech postdoctoral fellow Wang Na was a co-first author of the paper with Christopher Bosc from the University Grenoble Alpes and Sung Ryul Choi from the Paul Scherrer Institute.
Original paper – https://www.nature.com/articles/s41594-019-0241-y
Associate Professor Huang Hongda has also studied the molecular mechanism of VASH1, a paralog of VASH2. A related paper was published last month in Cell Research titled, “Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.” This paper revealed the structural basis of the detyrosination of tubulin by the VASH1-SVBP complex and the important role of VASH1-SVBP in cell division. Associate Professor Huang Hongda collaborated with Professor Xu Chao and his research team at the University of Science and Technology China (USTC). Postdoctoral fellow Wang Na was also the co-first author of this paper, along with Liao Shanhui from USTC and Girish Rajendraprasad from the Danish Cancer Society Research Center
Original paper – https://www.nature.com/articles/s41422-019-0187-y
X-ray data was collected at the Shanghai Synchroton Radiation Facility, and the Swiss Light Source at the Paul Scherrer Institute in Switzerland. The research was supported by grants from the National Key R&D Program of China, PIC-GIN (Photonic Imaging Center of GIN, University Grenoble Alpes-Inserm U1216, which is part of the ISdV core facility and certified by the IBiSA label), INSERM, CEA, University Grenoble Alpes, CNRS, La Ligue Contre le Cancer comité de l’Isère, fondation France Alzheimer, the Swiss National Science Foundation, the Shenzhen Government ‘Peacock Plan’ and the Thousand Young Talents Program.
Associate Professor Huang Hongda’s research group homepage: https://bio.sustc.edu.cn/?P=3817
DNA damage from various sources can adversely affect genome stability and cell viability. The most common DNA lesions, abasic sties, are generated by spontaneous base loss or base damage caused by ionizing radiation (IR), UV radiation (UV) and alkylating agents such as methyl methanesulfonate (MMS). Whilst these sites in double-stranded DNA are primarily repaired by a cellular mechanism known as the base excision repair (BER) pathway, their repair mechanism in single-stranded DNA is only just being uncovered.
Late last month, a research group led by Associate Professor Huang Hongda from the Department of Biology at Southern University of Science and Technology (SUSTech), had a paper published in the internationally renowned journal ‘Nucleic Acids Research’, entitled “Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK”. The paper sought to unveil the molecular basis of abasic sites sensing in ssDNA by yedK.
The abasic site is the most common type of DNA damage. It is estimated that each cell of the human body can produce 5,000 to 10,000 abasic sites per day. Abasic sites that are not repaired in time can hinder RNA polymerase transcription and DNA polymerase replication, which may lead to further mutations, genomic instability, and even cancer.
Previous studies have focused on damage repair of abasic sites on double-stranded DNA. Recently, the HMCES protein and its homologous protein yedK have been found to protect abasic sites on single-stranded DNA, thereby mediating high fidelity repair of abasic sites. However, the mechanism of action of HMCES and yedK proteins and how they mediate downstream repair pathways are unclear.
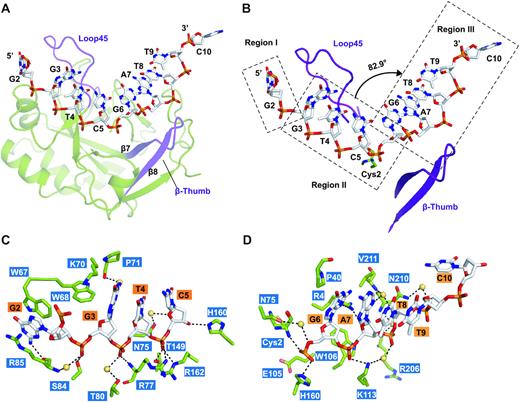
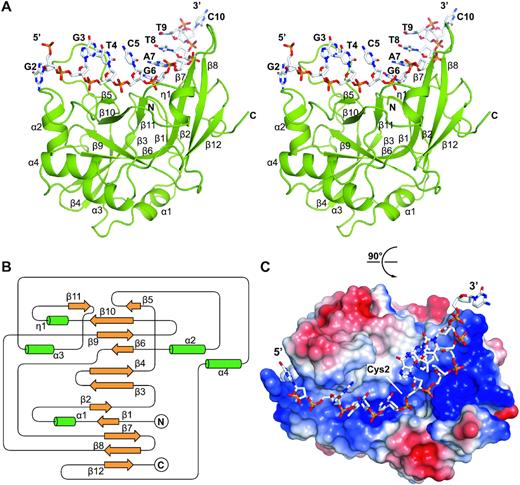
This study analyzed a series of high-resolution complex structures of yedK protein and single-stranded DNA, including the structure of yedK binding non-specific substrates (natural single-stranded DNA), the structure of binding specific substrates (tetrahydrofuran mimic of abasic site), and the structure of binding post-reaction products.
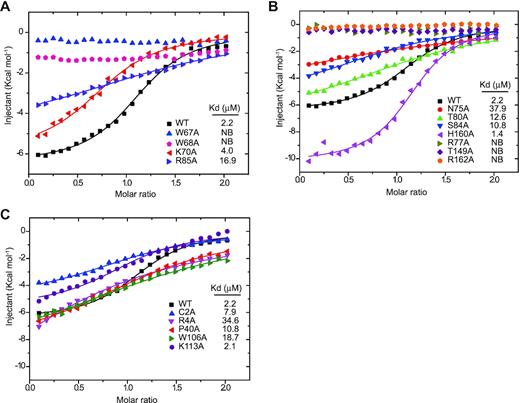
It is further revealed that yedK can specifically recognize the abasic site through the electrostatic repulsion between glutamic acid 105 and the phosphate group of single-stranded DNA; yedK nucleophilic attacks on the 1′-carbon atom of the abasic site in the single-stranded DNA through the thiol and a-amino groups of the cysteine residue in its active center;Finally, a thiazolidine ring is formed, which covalently crosslinks yedK protein and single-stranded DNA. The mechanism of yedK was further elucidated by in vitro biochemical and enzymatic assays.
The Department of Biology at SUSTech is the only correspondent unit of the article. Among them, the post-doctoral researchers Wang Na and Bao Hongyu are the co-first authors of the paper. Associate Professor Huang Hongda and post-doctoral researcher Wu Baixing are the correspondence authors of the paper.
All structural data of the study were collected at the beamlines BL19U/18U/17U of Shanghai Synchrotron Radiation Facility. The research team obtained funding from the National Key R&D Program of China, Shenzhen Government ‘Peacock Plan,’ Thousand Young Talents Program, and Chinese National Natural Science Foundation.
Original article – https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkz744/5555671
Proofread By
Photo ByDepartment of Biology