Small RNA (sRNA) are short RNA molecules, usually non-coding, involved with gene silencing and the post-transcriptional regulation of gene expression. It is an important eukaryotic gene regulation pathway, which plays a critical role in the development, gene silencing, and resistance of virus infections.
In the biogenesis of sRNA, Dicer family endonucleases are central to selecting the precursor sRNA (pre-sRNA), cutting the RNA to a defined length, and transferring one RNA strand to the downstream AGO protein, mediating the gene silencing. Since the 1990s, when it was discovered, it remains unclear how the Dicer proteins as a whole module perform the function of “molecular ruler” and “molecular scissors” to produce sRNA with defined lengths.
Therefore, a team of international researchers in this field have come together to collaborate in the research of gene silencing and resistance to virus infections. The team was led by Professor Jiamu Du of the Southern University of Science and Technology (SUSTech), Professor Steven E. Jacobsen of the University of California, Los Angeles (UCLA), and Professor Sisi Li of the School of Medicine at Shenzhen University.
Their research, entitled “Mechanism of siRNA production by a plant Dicer-RNA complex in dicing competent conformation,” was published in Science, one of the world’s top academic journals. It aims to make a milestone breakthrough in this field of sRNA.

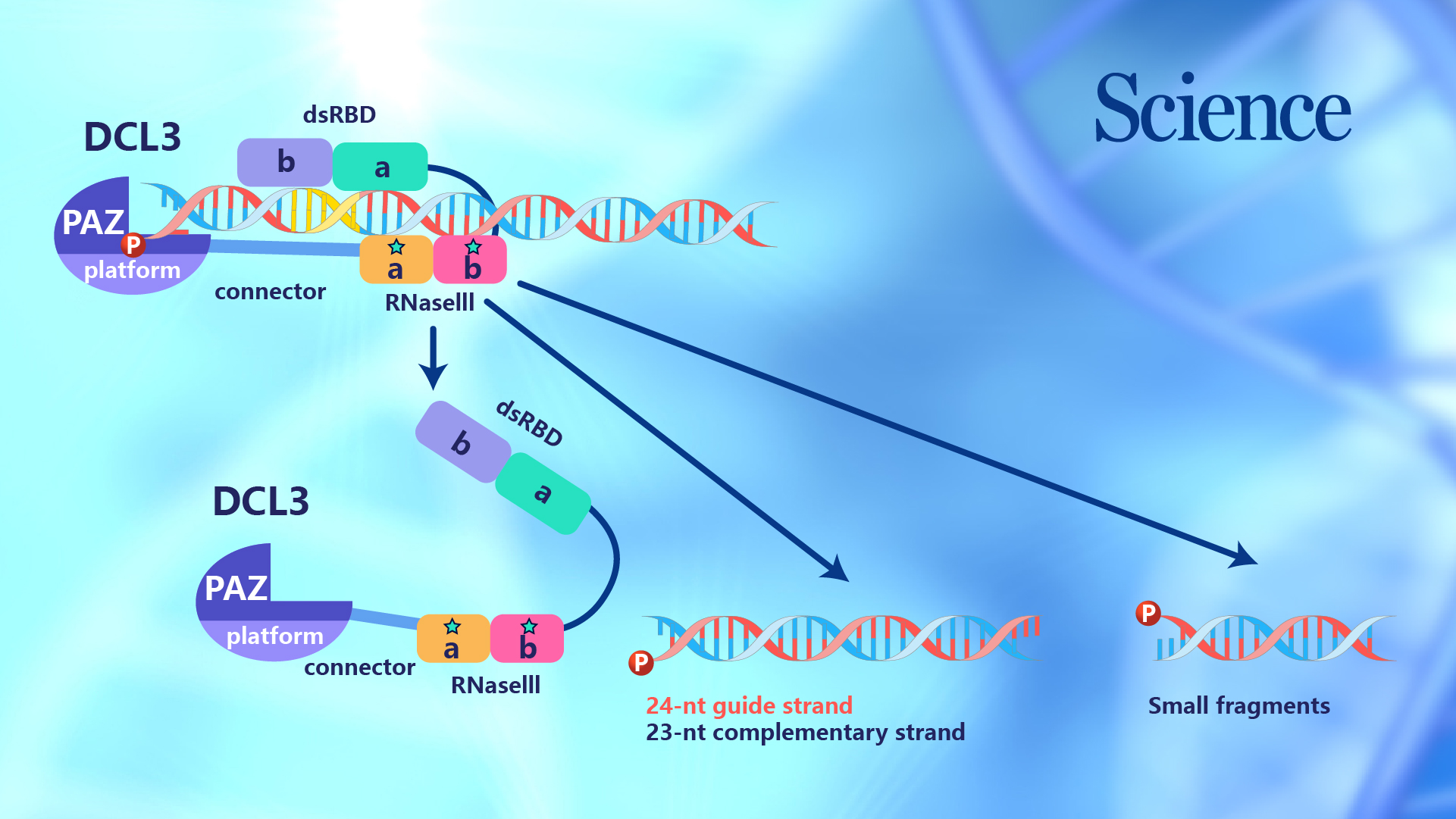
Figure 1. The overall structure of the DCL3-RNA complex
Deciphering small RNA through a plant-specific Dicer
In the sRNA biogenesis, Dicer is responsible for recognizing one end of the pre-sRNA and using itself as a ruler to measure and cut RNA. The researchers investigated the mechanism underlying the production of 24-nt siRNA by a plant-specific Dicer, Dicer Like 3 (DCL3). Based on the biochemical feature of DCL3, an RNA duplex with a guide strand possessing 5’-phosphorylated-A and a complementary strand possessing 1-nt 3’-overhang was designed and used to mimic the natural substrate of DCL3. In addition, the Ca2+ ions were added to the reaction system to mimic the Mg2+ ion but prevent dicing, which trapped the DCL3-RNA complex in a dicing-competent conformation.
Using the Cryo-Electron Microscopy Center at SUSTech, a complex structure of DCL3 with a 40-bp substrate RNA was determined. The structure readily showed that DCL3 separates the first base-pair of the RNA substrate by its platform-PAZ-connector cassette, thereby pushing the 5’-phosphorylated-A of the guide strand and the 3’-overhang of the complementary strand into two pockets to facilitate specific recognitions, respectively. This observation first observed the mechanism underlying the simultaneous recognition of both the 5’- and 3’-ends of the pre-siRNA.
Upon recognizing the RNA-termini by DCL3, the remaining part of the RNA was positioned into a binding channel formed by multiple domains of DCL3. DCL3 possesses multiple binding sites along with the RNA, and the binding extends to the RNaseIII domains. The two RNaseIII domains of DLC3 form a dimer and simultaneously cut the two strands of the RNA. In this case, one strand of RNA is cut between the 24th and 25th bases to produce a 24-nt product. Meanwhile, the other RNA strand is cut between the 23rd and 24th base to produce a 23-nt strand. As a result, this fully revealed the measuring mechanism of the DCL3. Because the downstream AGO4 only selects 24-nt siRNA, the structure also explained the basis for the strand-biased siRNA selection. The following biochemical and functional data further conformed to the termini-recognition and strand-biased cutting by DCL3.
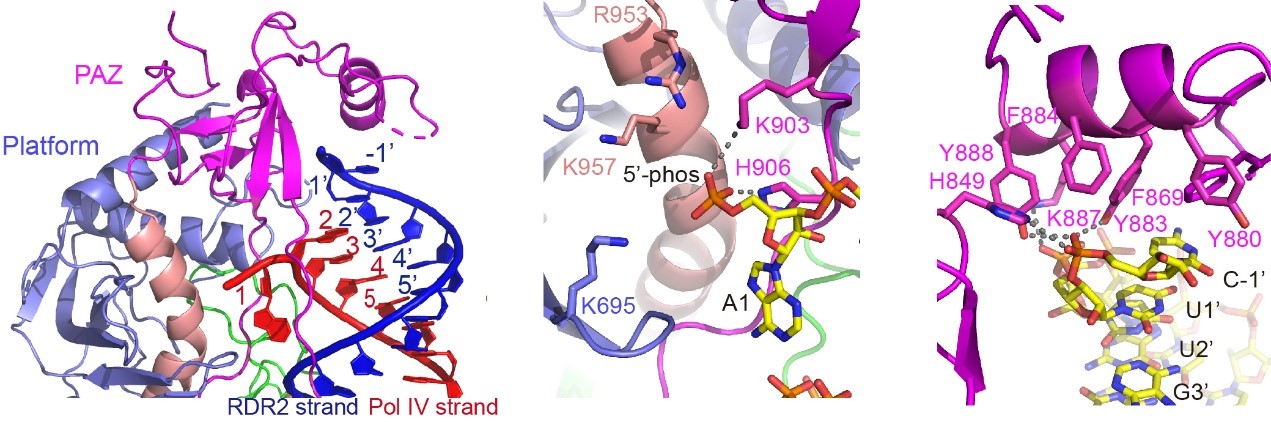
Figure 2. The recognition of RNA termini (left), 5’-phosphate (middle), and 3’-overhang (right) by DCL3
“Once we first saw the electron density map, we realized that this is the first dicing-competent conformation Dicer, and most of the dicing-related mechanism can be resolved immediately,” said Professor Jiamu Du from the Department of Biology at SUSTech.
This study has observed the dicing-competent conformation of a Dicer-RNA complex and also revealed the termini-specific, length-dependent, and strand-biased cutting. During the review process, one reviewer commented on the “landmark contribution” of the paper in this field.
Ms. Qian Wang, a Ph.D. student at SUSTech, Dr. Yan Xue from UCLA, and Dr. Laixing Zhang from Tsinghua University are the co-first authors of this paper. Professors Jiamu Du, Steven Jacobsen, and Sisi Li are the co-corresponding authors.
The Cryo-EM data was collected at the SUSTech Cryo-Electron Microscopy Center. Professor Maojun Yang from Tsinghua University and Professors Jixian Zhai and Zhe Wu from SUSTech helped with the data processing and functional insights.
This research was supported by the National Key R&D Program, Shenzhen Science Technology Programs, Guangdong Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes, and the Guangdong Innovation Research Team Fund.
Paper link: http://science.org/doi/10.1126/science.abl4546
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By