The three-dimensional structure of the genome is analyzed by Chromatin Conformation Capture technology by sequencing chimeric sequences from chromatin digestion and proximity ligation. Among them, the application of Hi-C and ChIA-PET technologies in plants has provided us with abundant information on the structure of plant genome, like Arabidopsis, rice, tomato, and maize. However, Hi-C and ChIA-PET technologies can only provide two-way interaction of chromosomal loci due to the limitation of Illumina paired-end short read.
The multi-way interaction between chromatin sites can only be inferred from the interaction information of two sites from multiple cells. Because of the cellular heterogeneity, this kind of inference is not accurate. In addition, the Hi-C and ChIA-PET techniques include PCR amplification steps and thus cannot retain the original epigenetic modification information on the DNA sequence. Other techniques for capturing chromatin multi-site interactions, such as SPRITE and ChIA-Drop, are challenging to operate and hard to be applied in plants.
Associate Professor Jixian Zhai’s research group from the Institute of Plant and Food Science of the School of Life Sciences at the Southern University of Science and Technology (SUSTech) recently applied Pore-C technology based on third-generation sequencing Nanopore platform to analyze the multi-way interactions and associated methylation modification in Arabidopsis genome.
Their study, entitled “Pore-C Simultaneously Captures Genome-wide Multi-way Chromatin Interaction and Associated DNA Methylation Status in Arabidopsis”, was published in Plant Biotechnology Journal.
Nanopore sequencing is a long-read, single-molecule sequencing technology that obtains the sequence context through the electric current change caused by the passing of nucleotide molecules through the pore. The length of the Nanopore read can generally reach 30kb, and various kinds of epigenetic modification information on the DNA molecules, including m5C, hm5C, and m6A, can be obtained together. In 2019, Ulahannan et al. developed Pore-C technology, using Nanopore to perform long-read sequencing of multi-fragment chimeric sequences formed after enzyme digestion and ligation of chromatin, and successfully analyzed the multi-site interaction of chromatins in mammalian cells.
In this study by Prof. Zhai’s team, the Pore-C technique was used in Arabidopsis for the first time to analyze the multi-site interactions in the plant genome. Compared with Hi-C, the Pore-C does not require biotin labeling, fragmentation, affinity purification, DNA purification, and other steps, greatly simplifying the experimental procedure (Figure 1A). The library construction process can be completed within three days. Good consistency was found between the Pore-C and Hi-C data and between the two Pore-C libraries, indicating the accuracy and repeatability of this technique (Figures 1B and 1C).
Compared with the Hi-C library, Pore-C obtained a comparable number of pair-wise interactions with fewer reads and sequenced bases, which proved that Pore-C is more efficient in detecting genomic interactions. In Arabidopsis, 44% Pore-C reads contained multiple genomic fragments (Figure 1D), similar to the performance of Pore-C in human cells (47%), and Pore-C’s detection efficiency of multi-site interaction is higher than SPRITE. Pore-C fragments with more detection sites can capture interactions among longer genomic distances, and this feature can be applied to assist genome assembly (Figure 1E).
The application of Pore-C in Arabidopsis confirms the speculation that there are interactions among several KNOT ENGAGED elements (KEE) in the Arabidopsis genome. More than 1145 Pore-C reads detected the interaction between three KEEs, and the contacts were confirmed by a statistical test. Among them, half of KEE multi-site interactions were associated with KEE3 and KEE4, indicating that KEE3 and KEE4 may be the core regions of KEE high-dimensional interactions (Figure 1F). In addition, multiple Pore-C reads detected the interactions between 4 KEEs (Figure 1G). Similarly, Pore-C detected multiple telomere interactions in Arabidopsis, which was consistent with the previous Hi-C data.
The methylation modification information in Pore-C data was also analyzed in this study. The researchers used the methylation modification analysis tool DeepSignal-Plant to analyze the distribution of CG, CHG, and CHH modification in Pore-C fragments. The methylation modification signals obtained were highly consistent with the traditional Whole Genome Bisulfite Sequencing (WGBS) results (Figure 1H). They found that interacting fragments with close spatial distances highly correlated in their CG methylation level, which was similar to the pattern found in mouse embryonic stem cells (mESC) in 2019 by Ren Bin’s group using the Methyl-HiC technique (Figure 1I).
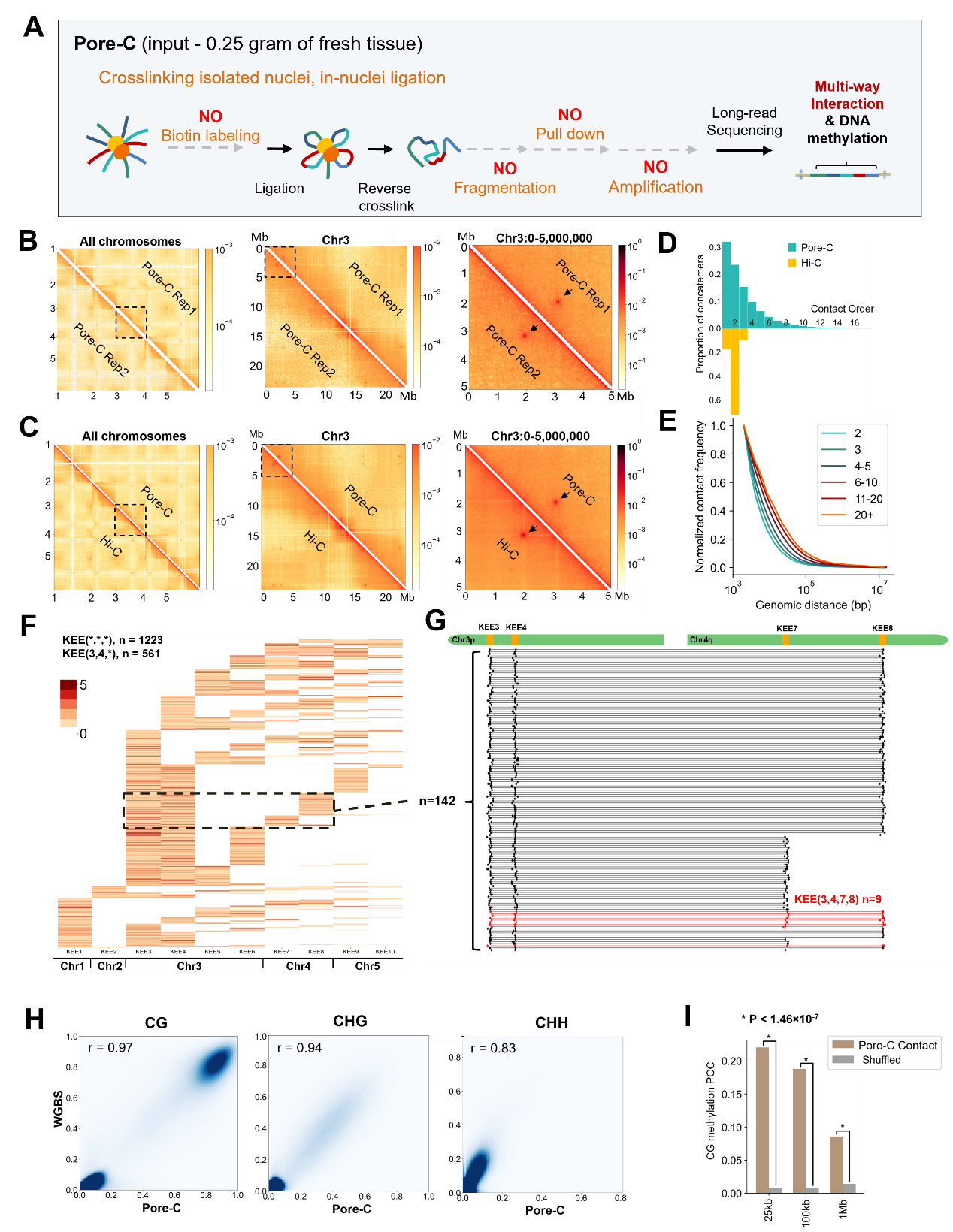
Figure 1. Pore-C captures multi-way contacts and associated DNA methylation in Arabidopsis. (A) Schematic of the Pore-C protocol. (B-C) The consistency between two Pore-C libraries between Hi-C and Pore-C, respectively. (D) Comparison of Contact Order distribution between previous Hi-C data and Pore-C data in Arabidopsis. (E) Contact frequency between different genomic distances of various order groups of Pore-C. (F) Combination information of reads related to at least three different KEEs. Each row of the heatmap represents one read. (G) Alignment locations of individual reads related to KEE3, KEE 4, KEE7, and KEE8. (H) The consistency of CG, CHG, CHH DNA methylation between Pore-C and WGBS data.
Research Assistant Prof. Yanping Long and Ph.D. student Zhuowen Li are the co-first authors of this paper. Associate Prof. Jixian Zhai is the corresponding author. Ph.D. students Yiming Yu, Zhijian Liu, Weipeng Mo, master’s student Hong Zhang, and Research Assistant Prof. Jinbu Jia also contributed to this study.
The research was supported by the National Key R&D Program of China Grant, the Program for Guangdong Introducing Innovative and Entrepreneurial Teams, and the Shenzhen Sci-Tech Fund.
Paper link: https://onlinelibrary.wiley.com/doi/10.1111/pbi.13811
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By