Polyadenylation is essential for eukaryotic mRNA maturation. The process of a gene using different polyA sites for generating different transcripts is called alternative polyadenylation (APA). APA involves in many post-transcriptional regulations, such as mRNA maturation, mRNA stability, cellular RNA decay, and RNA’s cellular localization.
The advances of next-generation sequencing resulted in the genome-wide mapping of polyA sites, which further revealed the features of polyA sites and the regulation of APA. However, our knowledge about ployA site usage and its dynamics at single-cell level is still limited.
Associate Professor Wenfei Jin from the School of Life Sciences at the Southern University of Science and Technology (SUSTech) led a research group that recently developed a single-cell polyA sequencing approach and used it to decipher the dynamics of polyA site usage at single-cell resolution.
Their research results were published in the high-impact academic journal Proceedings of the National Academy of Sciences (PNAS), entitled “Comprehensive mapping of alternative polyadenylation site usage and its dynamics at single-cell resolution”.
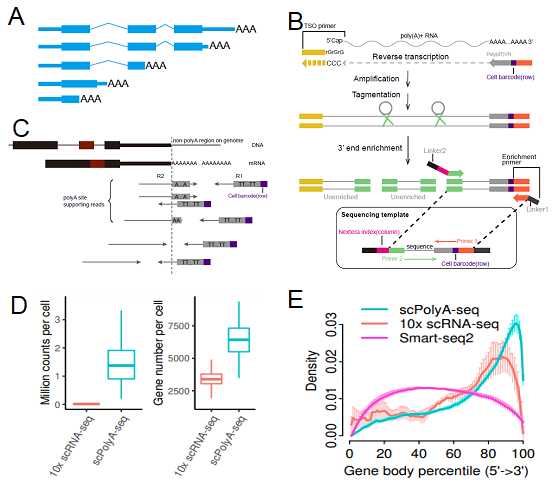
Figure 1. Summary of scPolyA-seq. (A) Scheme of a gene with multiple polyA sites. (B) Scheme of scPolyA-seq. (C) Scheme of polyA site supporting reads used for identification of polyA site. (D) scPolyA-seq generates much more reads and detects more genes for each cell compared with 10x scRNA-seq. (E) Reads from scPolyA-seq showed the sharpest peak at the 3’end of each gene, compared with 10x scRNA-seq and Smart-seq2.
In this study, single-cell polyadenylation sequencing (scPolyA-seq), a strand-specific approach for sequencing 3’ end of transcripts, and its data analyses pipeline were established to investigate the landscape of APA at single-cell level. By analyzing several cell lines, the researchers found many genes using multiple polyA sites in bulk data are prone to use only one polyA site in each single cell.
Interestingly, cell cycle genes were significantly enriched in genes with high variation in polyA site usage, as well as genes with the polyA site usage switch after cell synchronization. Genes showing cell cycle-associated polyA site usage switches were grouped into six clusters, with cell phase-specific functional categories enriched in each cluster. Deletion of one polyA site in MSL1 and SCCPDH results in slower and faster cell cycle progression, respectively, which supports that the polyA site usage switch played an important role in the cell cycle.
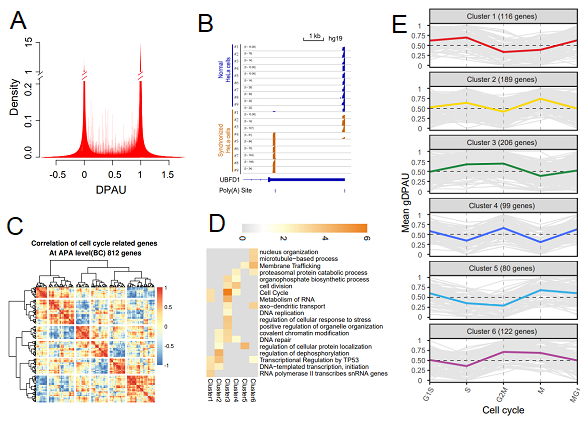
Figure 2. Feature of polyA site usage and its relationship with cell cycle. (A) Density of DPAU showing genes with two polyA sites across cells showed a bimodal curve, indicating these genes tend to only use one polyA site in a specific cell. (B) PolyA site usage switches of UBFD1 before and after synchronization of the HeLa cell. (C) Heatmap of correlation of gDPAU of each gene across five cell phases, resulting in genes clustered into six gene clusters. (D) GO enrichment analyses of each gene cluster. (E) Dynamics of gDPAU of each gene at each phase for the six gene clusters. Each line denotes one gene. Y-axis indicates this gene’s gDPAU at each phase. Colored lines are averaged gDPAU of each gene cluster.
This study was the first endeavour that systemically analysed polyA site usage dynamics in the cell cycle. Notably, they found multiple gene clusters with different dynamic patterns of polyA site usage, indicating that APA is an important layer for cell cycle regulation.
Assoc. Prof. Wenfei Jin at SUSTech and Prof. Ting Ni at Fudan University co-designed this study. Junliang Wang, a joint Ph.D. student at SUSTech and Zhengzhou University (ZZU), and Wei Chen, a Ph.D. student at Fudan University, are the co-first authors of this paper. Assoc. Prof. Wenfei Jin, Prof. Ting Ni, Research Asst. Prof. Ni Hong at SUSTech, and Prof. Yuanming Qi at ZZU are the co-corresponding authors.
This study was supported by the National Key R&D Program of China, National Natural Science Foundation of China (NSFC), Shenzhen Science and Technology Program, and Shenzhen Innovation Committee of Science and Technology.
Paper link: https://pubmed.ncbi.nlm.nih.gov/36454750
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo By