For many natural networked complex systems, evolution is their most striking feature. Exploring and understanding the underlying mechanism of evolution lies at the heart of many scientific areas, ranging from studying the origin and evolution of life and the ecological system to the emergence of intelligence through the brain neural network’s rewiring and formation of social communities and nation-states. It is the most fundamental aspect of scientific discoveries in evolving complex systems.
Complex networks, as a common form of networked complex system representation, are ubiquitous across a broad range of fields, from biology and ecology to social science, etc. It represents the complex relationships or interactions among the elements in a system and dictates its main functions. Therefore, knowing the evolution process of networked complex systems is essential to understanding the structure formation history of their corresponding complex networks.
However, complex networks typically have very rich mesoscopic structures, e.g., community structure, complex short-range loops, degree-degree correlation, and motifs. These structures usually exist simultaneously in a network, sharing complex interaction relationships, and work together to realize the basic functions of the system. Elucidating the relationships between these complex internal structures is generally difficult, let alone capturing the evolution mechanisms of the network. Existing research on the evolution of complex networks typically only focuses on one specific feature of real-world networks. For instance, the well-known preferential attachment mechanism can only explain the scale-free degree distribution, and even lead to contradictions with other common features, such as the community structure.
While it is difficult, if not impossible, to describe the evolution process of complex networks using concise rules by far, can we rely on machine learning techniques to restore their historical evolution trajectories? If it can be done, we can not only offer a basis for future research on the evolution mechanisms of complex networks, but also build data-driven models and generate new applications based on the restored evolutionary process. This would provide a giant leap in the research of evolving complex systems.
Associate Professor Yanqing Hu’s research group from the Department of Statistics and Data Science at the Southern University of Science and Technology (SUSTech) has recently published a paper reconstructing the formation history of networked complex systems with high precision and demonstrated the immense value in both scientific investigations and practical applications.
The paper, entitled “Reconstructing the evolution history of networked complex systems”, has been published in Nature Communications, and was additionally featured in the Editors’ Highlights of the journal under the Applied physics and mathematics category.
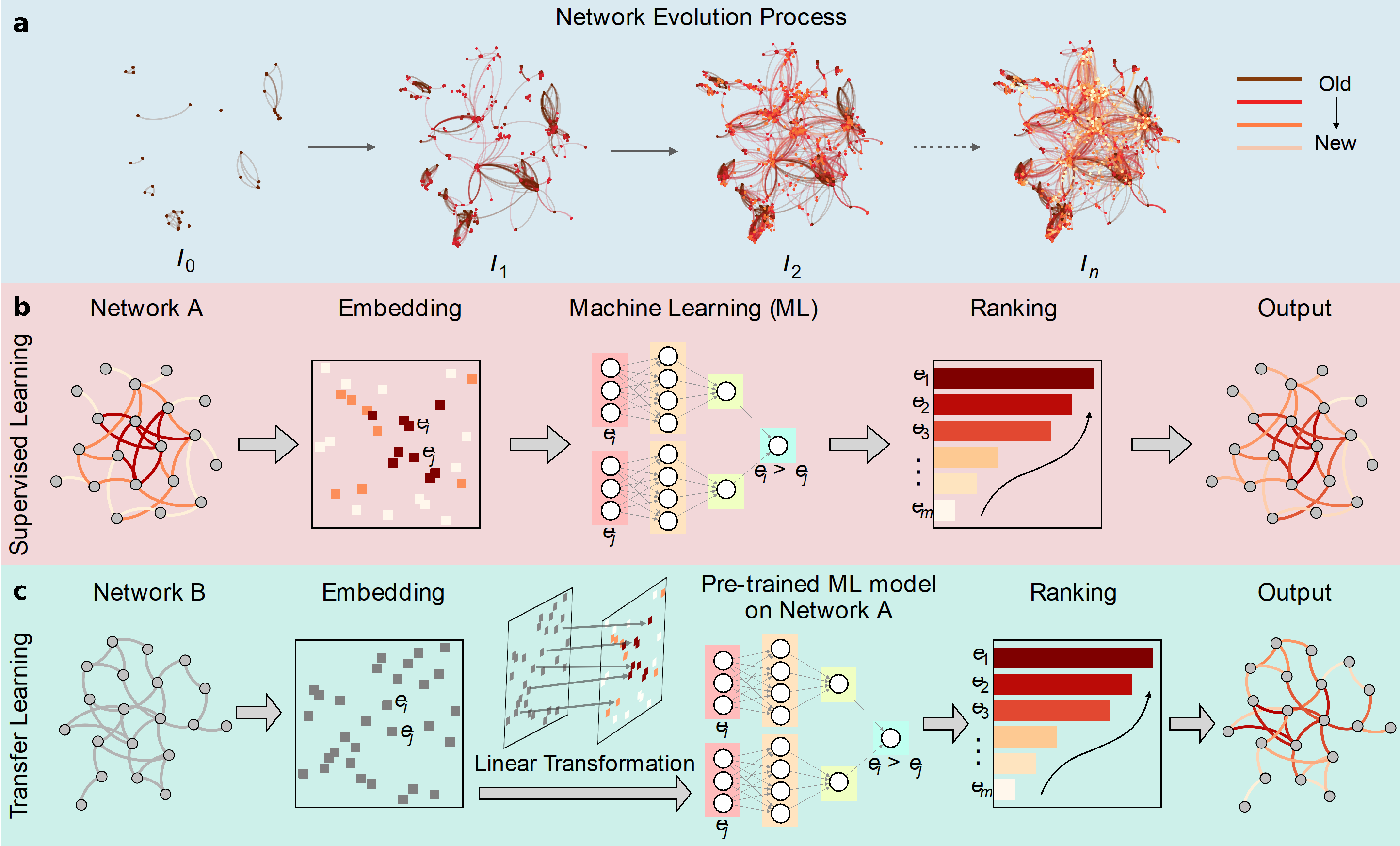
Figure 1. The network formation process and its restoration
Applying machine learning techniques (Figure 1), the research team demonstrated that the historical formation process of various networked complex systems can be extracted with high precision, including protein-protein interaction, ecology, and social network systems.
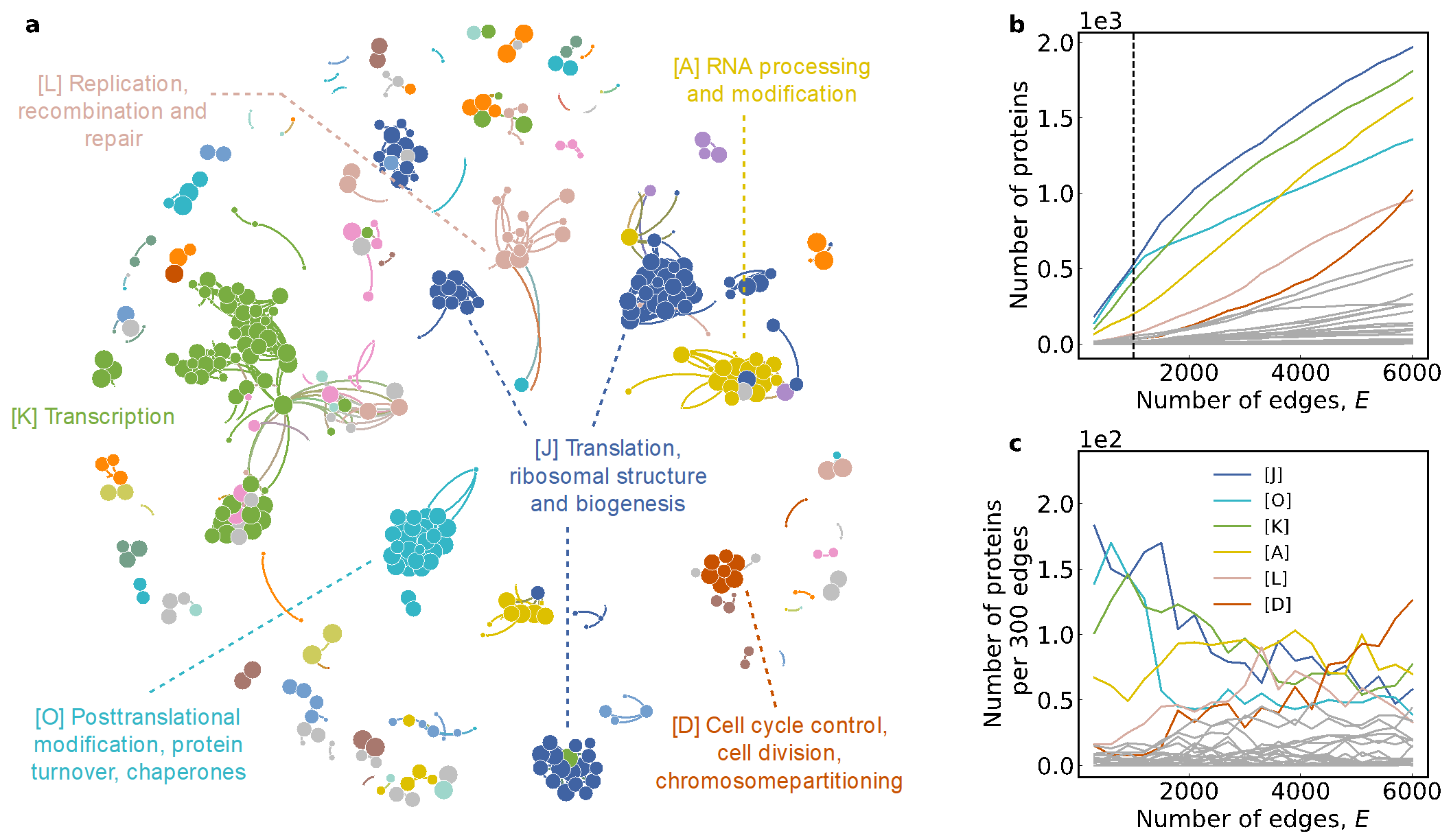
Figure 2. The restoration results help to interpret the evolution of protein-protein interaction networks
The recovered evolution process has demonstrated immense scientific value, particularly its functional properties in relation to the evolution dynamics. For instance, the formation trajectory of protein-protein interaction networks sheds light on the evolution process of living organisms at the molecular level (Figure 2). Infusing the temporal information sequence into link prediction mechanisms (Figure 3) can accelerate the process of drug development; the ecological network’s reconstruction can help us understand the history of co-evolution and predict future trends. In addition, the reconstructed results can reveal the key co-evolution features of network structures, such as preferential attachment, community structure, local clustering, and degree-degree correlation, that could not be explained collectively by previous theories.
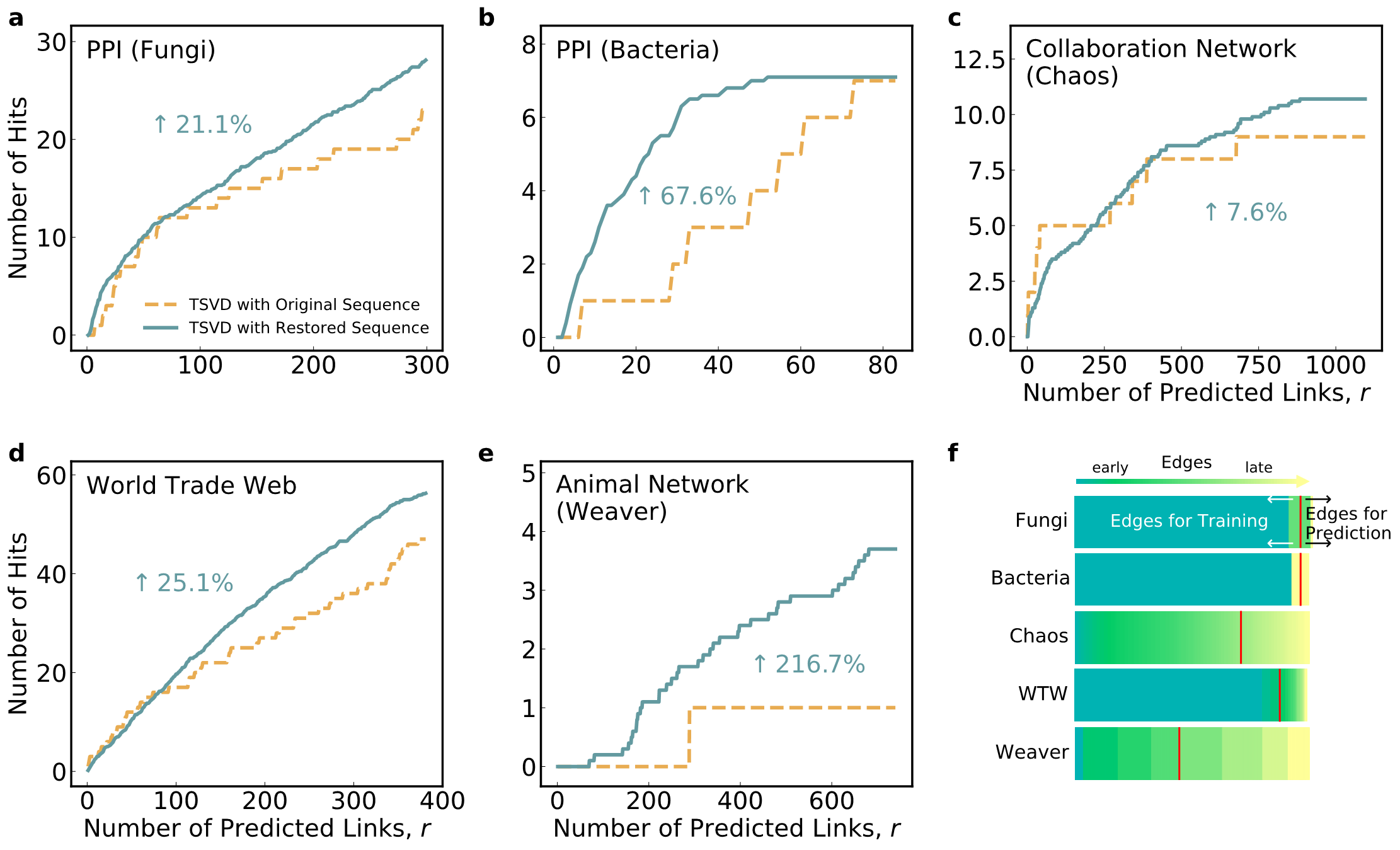
Figure 3. The restoration results improve the performance of link prediction
Intriguingly, the researchers discovered that for large networks, if the performance of the machine learning model is slightly better than a random guess on the pairwise order of links, reliable restoration of the overall network formation process can be achieved. This suggests that evolution history restoration is generally highly feasible on empirical networks.
Junya Wang, a Ph.D. student from Sun Yat-sen University, Yi-Jiao Zhang, a postdoctoral fellow from SUSTech, and Cong Xu, an instructor from SUSTech, are the co-first authors of the paper. Associate Professor Yanqing Hu is the corresponding author. Other contributors to this study included Ph.D. student Jiaze Li from Maastricht University, Dr. Jiachen Sun from Tencent Inc., Associate Professor Jiarong Xie from Beijing Normal University, Researcher Ling Feng from A*STAR in Singapore, and Professor Tianshou Zhou from Sun Yat-sen University. SUSTech is the only communication unit.
This work was supported by the National Natural Science Foundation of China.
Paper link: https://rdcu.be/dDm5w
Editors’ Highlights of Nature Communications: https://www.nature.com/ncomms/editorshighlights
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yingying XIA
Photo ByDepartment of Statistics and Data Science