Members of the genus Vibrio are widely distributed in marine and estuarine environments, and include numerous environmental opportunists as well as highly virulent pathogens, with profound implications for human health and seafood safety. The escalating crisis of antibiotic resistance has severely constrained therapeutic options against Vibrio infections, highlighting an urgent need for novel antibacterial mechanisms and technologies to overcome drug resistance and the lack of effective treatments.

A research team led by Associate Professor Yang Fu from the Department of Biochemistry and the Homeostatic Medicine Institute at the Southern University of Science and Technology (SUSTech), has made a major advance in understanding microbial competition mediated by antibacterial effectors of the type VI secretion system (T6SS). They identified TseVs, the first T6SS effector protein reported to exhibit highly species-specific antibacterial activity.
Their findings, entitled “A Vibrio-specific T6SS effector reshapes microbial competition by disrupting Vibrio bioenergetics,” have been published as the cover article in Cell Host & Microbe.
The team adopted an alternative approach focused on interspecies competition, investigating non-toxigenic environmental strains of Vibrio cholerae isolated from natural ecosystems. These strains lack the genes encoding cholera toxin (CT) and the toxin-coregulated pilus (TCP) and are thus non-pathogenic to humans. Remarkably, several of these non-pathogenic strains harbor active T6SSs and encode TseVs, a Vibrio-specific antibacterial effector. TseVs are translocated across bacterial membranes via the T6SS into susceptible Vibrio cells, where they induce potent bactericidal effects. This activity is effective in both host-associated contexts and in natural environments, enabling efficient clearance of sensitive Vibrio populations.
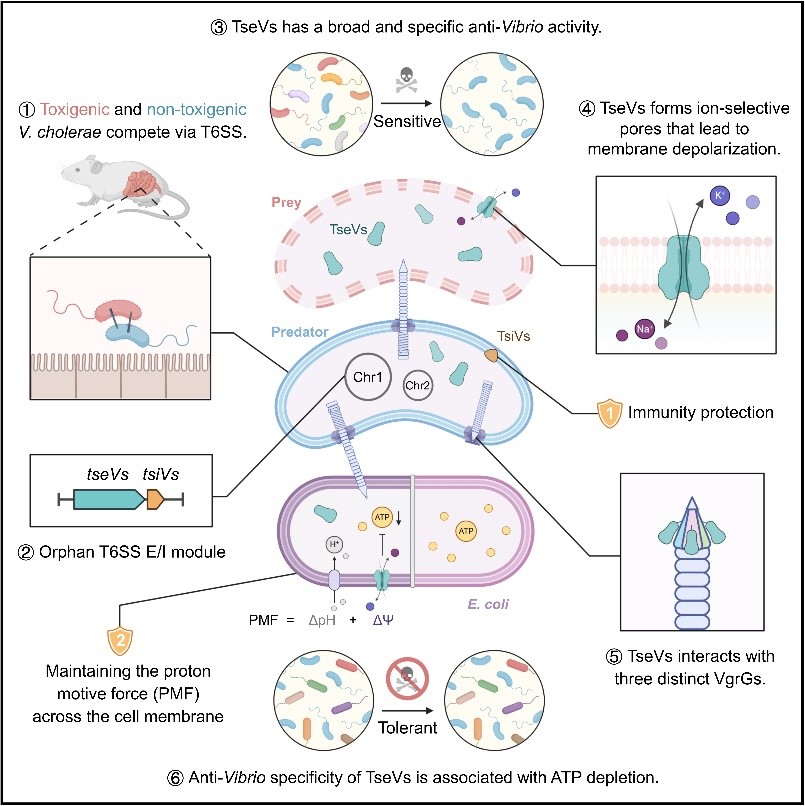
Figure 1. Species-specific antibacterial mechanism of the TseVs effector protein
Mechanistic investigations revealed that TseVs employ a glycine zipper–rich transmembrane helix domain to dimerize and form transmembrane pores spanning both the inner and outer membranes of target Vibrio cells. These pores permit Na⁺ influx and K⁺ efflux, selectively disrupting the ionic homeostasis of sensitive Vibrio, collapsing their ATP-generating capacity, and ultimately causing cell death. In contrast, non-Vibrio bacteria can restore their membrane potential via intrinsic ion homeostasis systems, thereby resisting TseVs-mediated killing without relying on the cognate immunity protein TsiVs.
Bioinformatic analyses showed that tseVs genes are predominantly found in non-pathogenic environmental Vibrio species and display signatures of evolutionary expansion in environmental samples from diverse geographic regions, underscoring their functional significance in shaping bacterial population dynamics.
This work expands the mechanistic understanding of T6SS-mediated microbial competition and establishes a compelling model for species-specific antibacterial strategies. TseVs combine strong taxonomic selectivity, high killing efficiency, low risk of resistance development, and minimal potential for horizontal gene transfer, positioning them as promising candidates for engineering into programmable antibacterial modules in synthetic biology. The findings also provide detailed ecological insights into the succession of pathogenic and non-pathogenic Vibrio communities and lay a robust theoretical foundation for the development of next-generation precision antimicrobial technologies.
Dr. Ming Liu from the Department of Biochemistry at SUSTech is the first author of the paper, and Associate Professor Yang Fu is the corresponding author. SUSTech is the first and sole corresponding institution.
Paper link: https://doi.org/10.1016/j.chom.2025.06.001
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
Proofread ByAdrian Cremin, Yuwen ZENG
Photo ByDepartment of Biochemistry